Downloads: 1
Diff of
/README.md
[000000]
..
[d15511]
Switch to side-by-side view
--- a +++ b/README.md @@ -0,0 +1,66 @@ +[](https://hippunfold.readthedocs.io/en/latest/?badge=latest) + + +<img align="right" width="200" src="https://github.com/khanlab/hippunfold/assets/25106300/0c16d33e-893a-4ac3-b127-21fa843823d5"> + +**Full Documentation:** [here](https://hippunfold.readthedocs.io/en/latest/?badge=latest) + +# Hippunfold + +This tool aims to automatically model the topological folding structure +of the human hippocampus, and computationally unfold it. +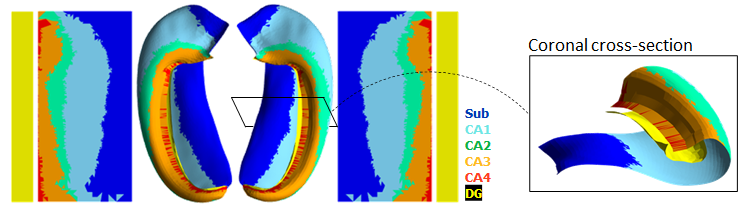 + +This is especially useful for: +- Visualization +- Topologically-constrained intersubject registration +- Parcellation (ie. registration to an unfolded atlas) +- Morphometry (eg. thickness, surface area, curvature, and gyrification measures) +- Quantitative mapping (eg. map your qT1 MRI data to a midthickness surface; extract laminar profiles perpendicular to this surface) + +## NEW: Version 1.3.x release + +Major changes include the addition of unfolded space registration to a reference atlas harmonized across seven ground-truth histology samples. This method allows shifting in unfolded space, providing even better intersubject alignment. + +*Note: this replaces the default workflow, however you can revert to the legacy workflow, disabling unfolded space registration, by setting `--atlas bigbrain` or `--no-unfolded-reg`* + +Read more in our [ manuscript](https://doi.org/10.7554/eLife.88404.3) + +Also the ability to specify a new **experimental** UNet model that is contrast-agnostic using [synthseg](https://github.com/BBillot/SynthSeg) and trained using more detailed segmentations. This generally produces more detailed results but has not been extensively tested yet. + +Note: Docker containers for version 1.3.x and above do not come pre-shipped with nnU-net models (and are accordingly more lightweight!) - models are downloaded automatically when running, but please see the FAQ for more information! + +## Workflow + +The overall workflow can be summarized in the following steps: + +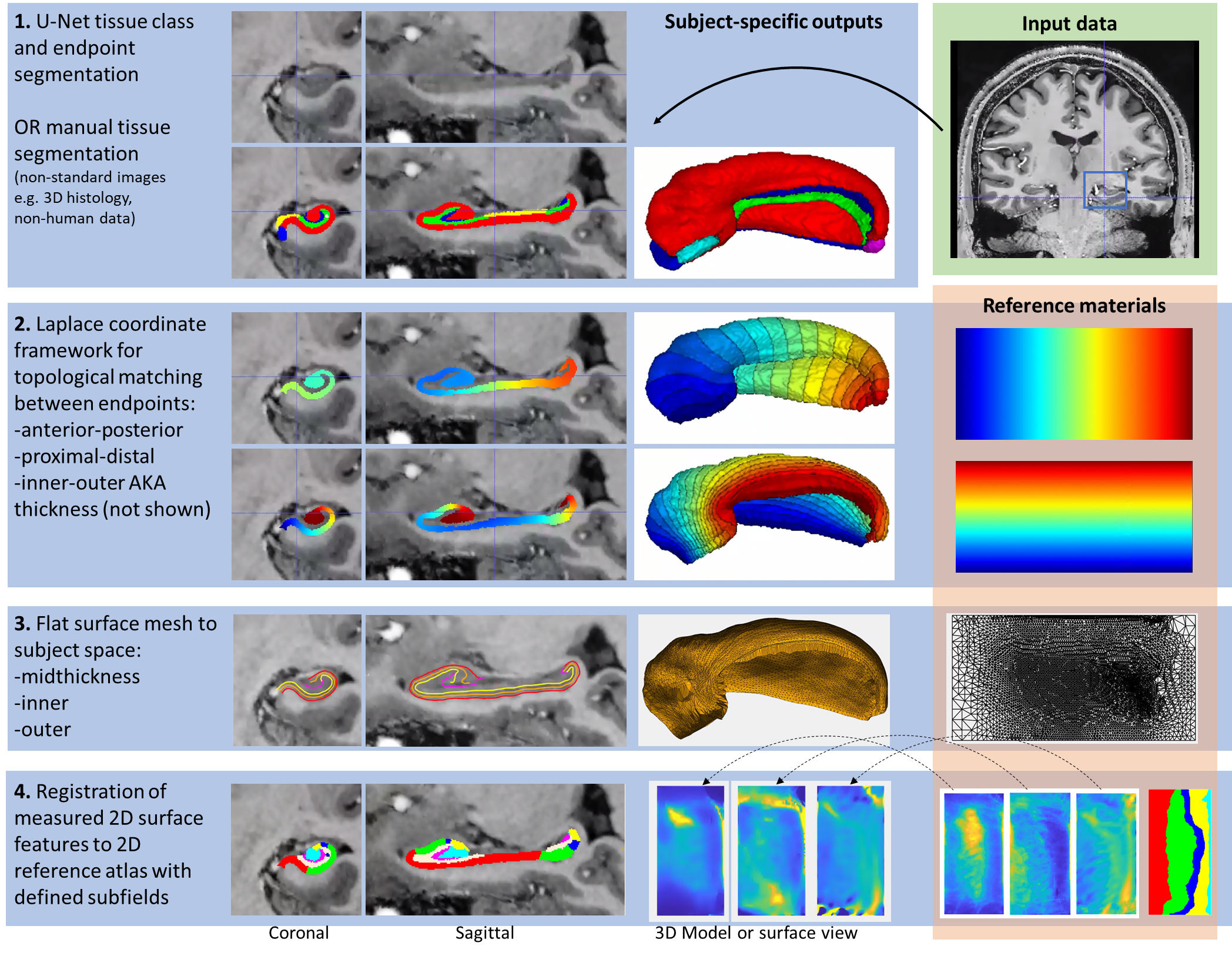 + +For more information, see +**Full Documentation:** [here](https://hippunfold.readthedocs.io/en/latest/?badge=latest) + +## Additional tools + +For plotting, mapping fMRI, DWI or other data, and manipulating surfaces, see [here](https://github.com/jordandekraker/hippunfold_toolbox) + +For statistical testing (spin tests) in unfolded space, see [here](https://github.com/Bradley-Karat/Hippo_Spin_Testing) + +## Publications + +### HippUnfold methods paper + +- DeKraker, J., Haast, R. A., Yousif, M. D., Karat, B., Lau, J. C., Köhler, S., & Khan, A. R. (2022). Automated hippocampal unfolding for morphometry and subfield segmentation with HippUnfold. Elife, 11, e77945. [link](https://doi.org/10.7554/eLife.77945) + - **Please cite this if you use any version of HippUnfold)** + +### Unfolded space registration and multihist7 atlas +- DeKraker Jordan, Palomero-Gallagher Nicola, Kedo Olga, Ladbon-Bernasconi Neda, Muenzing Sascha E.A., Axer Markus, Amunts Katrin, Khan Ali R., Bernhardt Boris, Evans Alan C. (2023) Evaluation of surface-based hippocampal registration using ground-truth subfield definitions eLife 12:RP88404 [link](https://doi.org/10.7554/eLife.88404.3) + - **Please cite this if you use HippUnfold version >= 1.3.0)** + +### Commentary on surface-based hippocampal segmentation +- DeKraker J, Köhler S, Khan AR. Surface-based hippocampal subfield segmentation. Trends Neurosci. 2021 Nov;44(11):856-863. doi: 10.1016/j.tins.2021.06.005. Epub 2021 Jul 22. PMID: 34304910. [link](https://pubmed.ncbi.nlm.nih.gov/34304910/) + +### Related papers + +- DeKraker J, Ferko KM, Lau JC, Köhler S, Khan AR. Unfolding the hippocampus: An intrinsic coordinate system for subfield segmentations and quantitative mapping. Neuroimage. 2018 Feb 15;167:408-418. doi: 10.1016/j.neuroimage.2017.11.054. Epub 2017 Nov 23. PMID: 29175494. [link](https://pubmed.ncbi.nlm.nih.gov/29175494/) +- DeKraker J, Lau JC, Ferko KM, Khan AR, Köhler S. Hippocampal subfields revealed through unfolding and unsupervised clustering of laminar and morphological features in 3D BigBrain. Neuroimage. 2020 Feb 1;206:116328. doi: 10.1016/j.neuroimage.2019.116328. Epub 2019 Nov 1. PMID: 31682982. [link](https://pubmed.ncbi.nlm.nih.gov/31682982/) +- Karat BG, DeKraker J, Hussain U, Köhler S, Khan AR. Mapping the macrostructure and microstructure of the in vivo human hippocampus using diffusion MRI. Hum Brain Mapp. 2023 Nov;44(16):5485-5503. Epub 2023 Aug 24. PMID: 37615057; PMCID: PMC10543110.[link](https://doi.org/10.1002/hbm.26461)

