Downloads: 1
Switch to side-by-side view
--- a/README.md +++ b/README.md @@ -1,66 +1,66 @@ -# Brain segmentation - -This is a source code for the deep learning segmentation used in the paper [Association of genomic subtypes of lower-grade gliomas with shape features automatically extracted by a deep learning algorithm.](https://doi.org/10.1016/j.compbiomed.2019.05.002) -It employs a U-Net like network for skull stripping and FLAIR abnormality segmentation. -This repository contains a set of functions for data preprocessing (MatLab), training and inference (Python). -Weights for trained models are provided and can be used for deep learning based skull stripping or fine-tuning on a different dataset. -If you use our model or weights, please cite: - -``` -@article{buda2019association, - title={Association of genomic subtypes of lower-grade gliomas with shape features automatically extracted by a deep learning algorithm}, - author={Buda, Mateusz and Saha, Ashirbani and Mazurowski, Maciej A}, - journal={Computers in Biology and Medicine}, - volume={109}, - year={2019}, - publisher={Elsevier}, - doi={10.1016/j.compbiomed.2019.05.002} -} -``` - -Developed by [mateuszbuda](https://github.com/mateuszbuda). - -The repository is divided into two folders. -One for skull stripping and one for FLAIR abnormality segmentation. -They are based on the same model architecture but can be used separately. - -## Prerequisites - -- MatLab 2016b for pre-processing -- Python 2 with dependencies listed in the `requirements.txt` file -``` -sudo pip install -r requirements.txt -``` - -## Results - -Below we show qualitative results for the average and median case. -Blue outline corresponds to ground truth and red to the final automatic segmentation output. -Images show FLAIR modality after preprocessing and skull stripping. - -| Average Case | Median Case | -|:----------:|:---------:| -||| - -The distribution of Dice similarity coefficient (DSC) for the whole dataset of 110 cases used in our study. - - - -The red vertical line corresponds to mean DSC (83.60%) and the green one to median DSC (87.33%). - -## Trained weights - -To download trained weights use `download_weights.sh` script located in both skull stripping or flair segmentation folder. -It downloads *.h5 file with weights corresponding to training log shown in each task specific folder and responsible for the results reported there. - -## U-Net architecture - -The figure below shows a U-Net architecture implemented in this repository. - - - -## Data - - - -[kaggle.com/mateuszbuda/lgg-mri-segmentation](https://www.kaggle.com/mateuszbuda/lgg-mri-segmentation) +# Brain segmentation + +This is a source code for the deep learning segmentation used in the paper [Association of genomic subtypes of lower-grade gliomas with shape features automatically extracted by a deep learning algorithm.](https://doi.org/10.1016/j.compbiomed.2019.05.002) +It employs a U-Net like network for skull stripping and FLAIR abnormality segmentation. +This repository contains a set of functions for data preprocessing (MatLab), training and inference (Python). +Weights for trained models are provided and can be used for deep learning based skull stripping or fine-tuning on a different dataset. +If you use our model or weights, please cite: + +``` +@article{buda2019association, + title={Association of genomic subtypes of lower-grade gliomas with shape features automatically extracted by a deep learning algorithm}, + author={Buda, Mateusz and Saha, Ashirbani and Mazurowski, Maciej A}, + journal={Computers in Biology and Medicine}, + volume={109}, + year={2019}, + publisher={Elsevier}, + doi={10.1016/j.compbiomed.2019.05.002} +} +``` + +Developed by [mateuszbuda](https://github.com/mateuszbuda). + +The repository is divided into two folders. +One for skull stripping and one for FLAIR abnormality segmentation. +They are based on the same model architecture but can be used separately. + +## Prerequisites + +- MatLab 2016b for pre-processing +- Python 2 with dependencies listed in the `requirements.txt` file +``` +sudo pip install -r requirements.txt +``` + +## Results + +Below we show qualitative results for the average and median case. +Blue outline corresponds to ground truth and red to the final automatic segmentation output. +Images show FLAIR modality after preprocessing and skull stripping. + +| Average Case | Median Case | +|:----------:|:---------:| +||| + +The distribution of Dice similarity coefficient (DSC) for the whole dataset of 110 cases used in our study. + +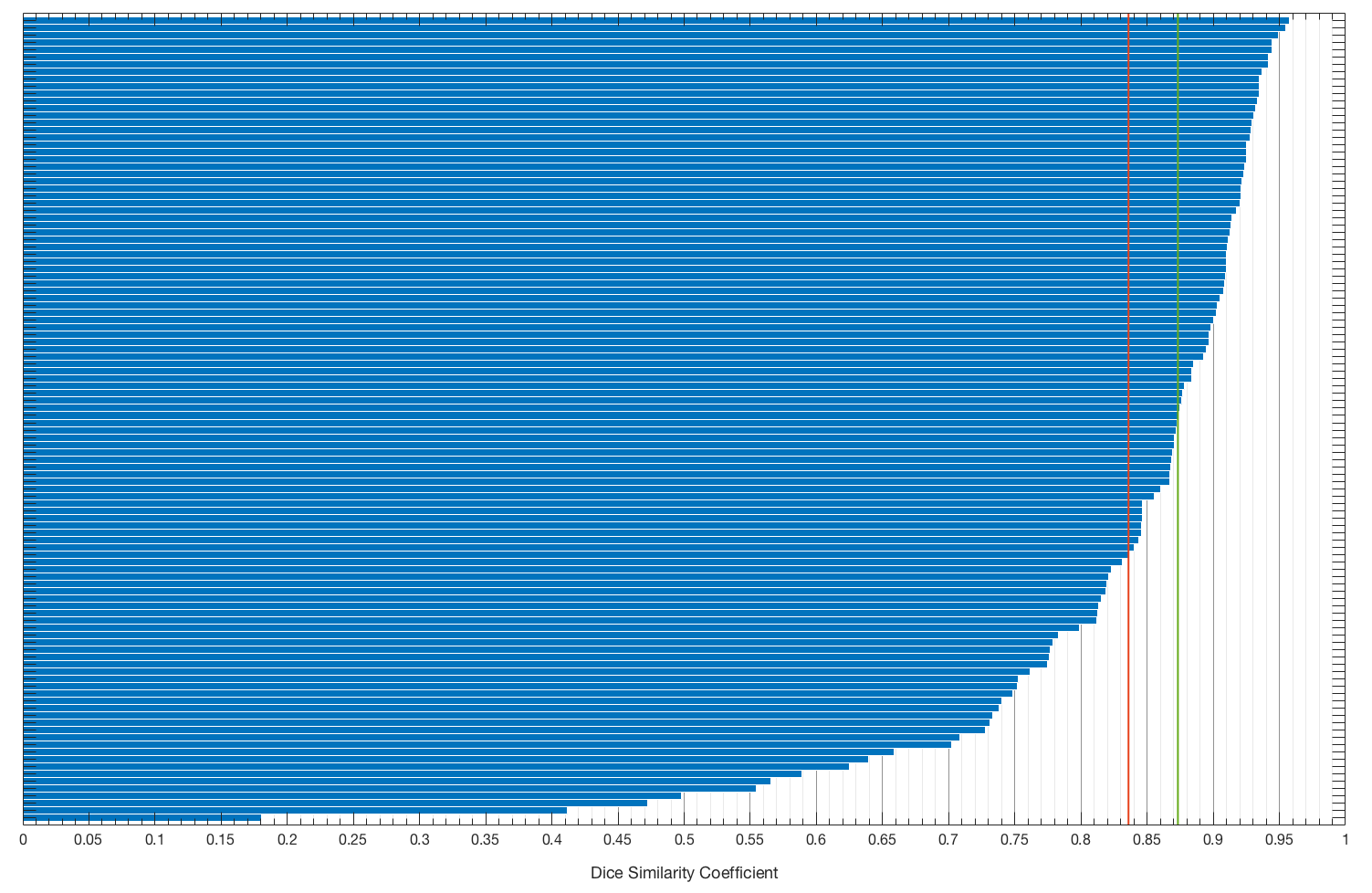 + +The red vertical line corresponds to mean DSC (83.60%) and the green one to median DSC (87.33%). + +## Trained weights + +To download trained weights use `download_weights.sh` script located in both skull stripping or flair segmentation folder. +It downloads *.h5 file with weights corresponding to training log shown in each task specific folder and responsible for the results reported there. + +## U-Net architecture + +The figure below shows a U-Net architecture implemented in this repository. + + + +## Data + + + +[kaggle.com/mateuszbuda/lgg-mri-segmentation](https://www.kaggle.com/mateuszbuda/lgg-mri-segmentation)

