Downloads: 1
Diff of
/README.md
[000000]
..
[69507b]
Switch to unified view
| a | b/README.md | ||
|---|---|---|---|
| 1 | ## Coronavirus disease 2019 (COVID-19) X-Ray Scanner for Diagnosis Triage |
||
| 2 | <img src="images/sample.png"> |
||
| 3 | |||
| 4 | This model is meant to help triage patients (prioritize certain patients for testing, quarantine, and medical attention) |
||
| 5 | that require diagnosis for COVID-19. This model is not meant to diagnose COVID-19. |
||
| 6 | |||
| 7 | It takes both PA and AP X-ray images (in DICOM format) as inputs and outputs a prediction for each image from one |
||
| 8 | of three labels (covid, opacity, nofinding). Those predicted with the labels "covid" and "opacity", as well as additional risk factors (Age, X-ray View Position) taken from the metadata, are flagged by the model for priority action. |
||
| 9 | |||
| 10 | The model analyzes each image for features predominant to COVID-19 images and for features present in images with Lung Opacity. If the model does not detect these features from the images, it returns a "nofinding" prediction. A "nofinding" prediction does not mean the patient is not sick for other diseases. The "covid" and "opacity" labels are not mutually exclusive. Mild and severe cases of COVID-19 exhibit lung opacity in images (which is also an idicator of Pneumonia), so the ideal worst case for the model is that an image from a patient who truly has COVID-19 symptoms is misclassified as "opacity". Therefore, the patient will still get priority action. This mitigates the current lack of COVID-19 X-ray data publicly available and the problem of false-negatives for binary classifiers. |
||
| 11 | |||
| 12 | **For example**, a 54-year old patient predicted to have lung opacity is flagged as higher priority for action by the model over a 20-year old patient predicted to have symptoms of COVID-19. Both are still flagged as higher priority for COVID-19 testing compared to those who were predicted to have no findings. |
||
| 13 | |||
| 14 | **Demo Video:** |
||
| 15 | |||
| 16 | [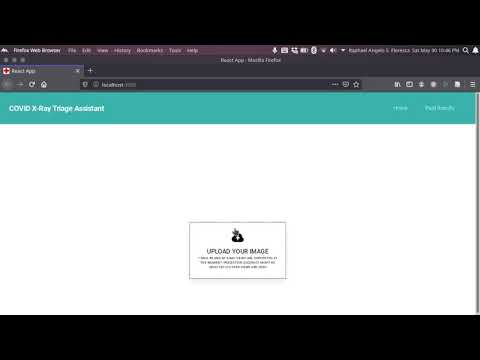](https://www.youtube.com/watch?v=NSQoiGwCB80) |
||
| 17 | |||
| 18 | ## Features |
||
| 19 | |||
| 20 | <img src="images/app%20screenshot.png"> |
||
| 21 | |||
| 22 | 1. Classifier (A.I. model) <br/> |
||
| 23 | 2. Flagging Risk Factors <br/> |
||
| 24 | 3. Visualization for Classifier explainability (Grad-CAM) <br/> |
||
| 25 | |||
| 26 | ## Specifications |
||
| 27 | Architecture: Resnet 34 <br/> |
||
| 28 | Training dataset: 26,000 images with weighted resampling <br/> |
||
| 29 | Training dataset size before Resampling: <br/> |
||
| 30 | |||
| 31 | 1. "covid" - 186 <br/> |
||
| 32 | 2. "opacity" - 5801 <br/> |
||
| 33 | 3. "nofinding" - 19884 <br/> |
||
| 34 | *Note: "nofinding" images include both healthy and non-healthy lungs that do not exhibit opacity |
||
| 35 | |||
| 36 | The model was trained on both AP and PA Chest X-rays. |
||
| 37 | |||
| 38 | Area under the Receiver Operator Characteristic (AUROC) was the chief metric used to determine model performance. It was calculated with a one-vs-all approach. |
||
| 39 | |||
| 40 | AUROC for "covid", "opacity", "nofinding" were at 99.97%, 92.64%, and 92.73%, respectively. |
||
| 41 | |||
| 42 | ## Resources |
||
| 43 | Updated Model (Use with Pytorch; .pth) : https://www.dropbox.com/s/o27w0dik8hdjaab/corona_resnet34.pth?dl=0 <br/> |
||
| 44 | |||
| 45 | Datasets used: 1. https://github.com/ieee8023/covid-chestxray-dataset <br/> |
||
| 46 | |||
| 47 | 2. https://www.kaggle.com/c/rsna-pneumonia-detection-challenge <br/> |
||
| 48 | |||
| 49 | Test_results: check the csv file in the repository <br/> |
||
| 50 | |||
| 51 | Colab_notebook (online version): check repository (Notice: <strong>NOT A DIAGNOSTIC TOOL</strong>) <br/> |
||
| 52 | |||
| 53 | ## Installation (Application version) |
||
| 54 | |||
| 55 | I: Install [Docker](https://docs.docker.com/get-docker/) appropriate for your computer |
||
| 56 | |||
| 57 | II. In the terminal: |
||
| 58 | |||
| 59 | 1. Clone the repository |
||
| 60 | 2. Set the current directory to the root of this repository |
||
| 61 | 3. Run ``` docker-compose up -d ``` |
||
| 62 | 4. After the setup is finished, it should open a new browser where the app is located. |
||
| 63 | 5. To stop the application, run ``` docker-compose stop ``` |
||
| 64 | |||
| 65 | ## (Previous version) Coronavirus disease 2019 (COVID-19) X-Ray Scanner |
||
| 66 | For the first iteration of the model, I built a CNN neural network that classifies a given Chest X-ray as positive for pneumonia caused by COVID-19 or not. This model is originally meant to demonstrate a proof-of-concept. The model was trained, and accepts, Posteroanterior views only. |
||
| 67 | |||
| 68 | Rationale/More information: https://towardsdatascience.com/using-deep-learning-to-detect-ncov-19-from-x-ray-images-1a89701d1acd<br/> |
||
| 69 | |||
| 70 | ## Acknowledgements |
||
| 71 | |||
| 72 | We thank Dr. Jianshu Weng, Mr. Najib Ninaba and their organisation, [AI Singapore](http://www.aisingapore.org/) (AISG), for their generous support in providing the infrastructure to train the latest iteration of the model. |
||
| 73 | |||
| 74 | Thank you to Ivan Leo for developing the application. Thank you to Chris, Raphael and Sunny for assisting in the application debugging. |
||
| 75 | |||
| 76 | ## LICENSE |
||
| 77 | <a rel="license" href="https://opensource.org/licenses/MIT"><img alt="Creative Commons Licence" style="border-width:0" src="https://upload.wikimedia.org/wikipedia/commons/thumb/0/0c/MIT_logo.svg/220px-MIT_logo.svg.png" /></a> |

