Downloads: 1
Switch to side-by-side view
--- a/README.md +++ b/README.md @@ -1,64 +1,64 @@ -# Pytorch implementation of 3D UNet - -This implementation is based on the orginial 3D UNet paper and adapted to be used for MRI or CT image segmentation task -> Link to the paper: [https://arxiv.org/pdf/1606.06650v1.pdf](https://arxiv.org/pdf/1606.06650v1.pdf) - -## Model Architecture - -The model architecture follows an encoder-decoder design which requires the input to be divisible by 16 due to its downsampling rate in the analysis path. - - - -## Dataset - -The Dataset class used for training the network is specially adapted to be used for the **Medical Segmentation Decathlon challenge**. - -This dataset contains several segmentation tasks on various organs including **Liver Tumours, Brain Tumours, Hippocampus, Lung Tumours, Prostate, Cardiac, -Pancreas Tumour, Colon Cancer, Hepatic Vessels and Spleen segmentation**. - -- Please also note that in the case which the task contain more than 2 classes (1: for foreground, 0: for background), you will need to modify the output -of the model to reshape it to the size of the groundtruth mask in train.py file. - -> The link to the dataset: [http://medicaldecathlon.com/](http://medicaldecathlon.com/) - -- The Dataset class uses Monai package for reading MRI or CT and also applying augmentations on them in the transform.py file. You can modify the applied -transformation in this file according to your preferences. - -## Configure the network - -All the configurations and hyperparameters are set in the config.py file. -Please note that you need to change the path to the dataset directory in the config.py file before running the model. - -**Parameters:** - -- DATASET_PATH -> the directory path to dataset .tar files - -- TASK_ID -> specifies the the segmentation task ID (see the dict below for hints) - -- IN_CHANNELS -> number of input channels - -- NUM_CLASSES -> specifies the number of output channels for dispirate classes - -- BACKGROUND_AS_CLASS -> if True, the model treats background as a class - -- TRAIN_VAL_TEST_SPLIT -> delineates the ratios in which the dataset shoud be splitted. The length of the array should be 3. - -- TRAINING_EPOCH -> number of training epochs - -- VAL_BATCH_SIZE -> specifies the batch size of the training DataLoader - -- TEST_BATCH_SIZE -> specifies the batch size of the test DataLoader - -- TRAIN_CUDA -> if True, moves the model and inference onto GPU - -- BCE_WEIGHTS -> the class weights for the Binary Cross Entropy loss - -## Training - -After configure config.py, you can start to train by running - -`python train.py` - -We also employ tensorboard to visualize the training process. - -`tensorboard --logdir=runs/` +# Pytorch implementation of 3D UNet + +This implementation is based on the orginial 3D UNet paper and adapted to be used for MRI or CT image segmentation task +> Link to the paper: [https://arxiv.org/pdf/1606.06650v1.pdf](https://arxiv.org/pdf/1606.06650v1.pdf) + +## Model Architecture + +The model architecture follows an encoder-decoder design which requires the input to be divisible by 16 due to its downsampling rate in the analysis path. + +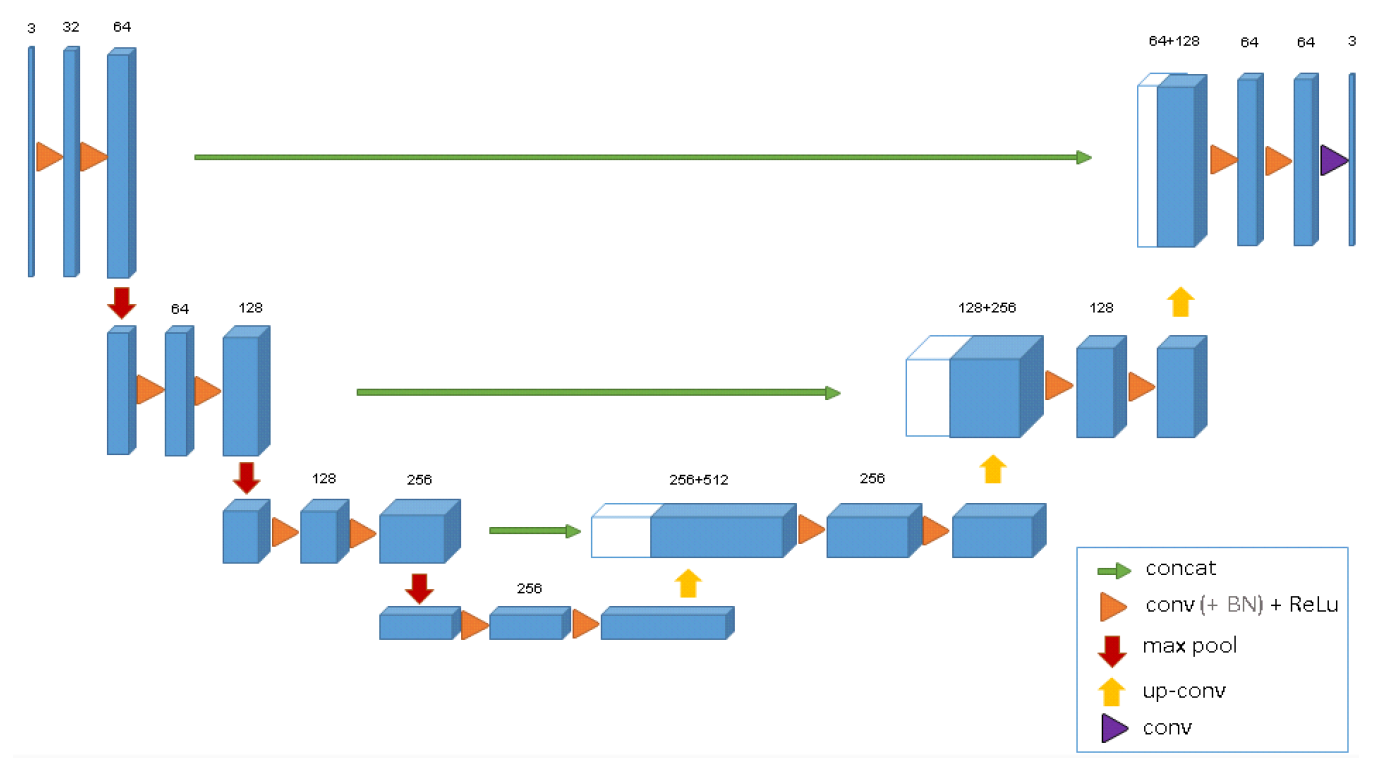 + +## Dataset + +The Dataset class used for training the network is specially adapted to be used for the **Medical Segmentation Decathlon challenge**. + +This dataset contains several segmentation tasks on various organs including **Liver Tumours, Brain Tumours, Hippocampus, Lung Tumours, Prostate, Cardiac, +Pancreas Tumour, Colon Cancer, Hepatic Vessels and Spleen segmentation**. + +- Please also note that in the case which the task contain more than 2 classes (1: for foreground, 0: for background), you will need to modify the output +of the model to reshape it to the size of the groundtruth mask in train.py file. + +The link to the dataset: [http://medicaldecathlon.com/](http://medicaldecathlon.com/) + +- The Dataset class uses Monai package for reading MRI or CT and also applying augmentations on them in the transform.py file. You can modify the applied +transformation in this file according to your preferences. + +## Configure the network + +All the configurations and hyperparameters are set in the config.py file. +Please note that you need to change the path to the dataset directory in the config.py file before running the model. + +**Parameters:** + +- DATASET_PATH -> the directory path to dataset .tar files + +- TASK_ID -> specifies the the segmentation task ID (see the dict below for hints) + +- IN_CHANNELS -> number of input channels + +- NUM_CLASSES -> specifies the number of output channels for dispirate classes + +- BACKGROUND_AS_CLASS -> if True, the model treats background as a class + +- TRAIN_VAL_TEST_SPLIT -> delineates the ratios in which the dataset shoud be splitted. The length of the array should be 3. + +- TRAINING_EPOCH -> number of training epochs + +- VAL_BATCH_SIZE -> specifies the batch size of the training DataLoader + +- TEST_BATCH_SIZE -> specifies the batch size of the test DataLoader + +- TRAIN_CUDA -> if True, moves the model and inference onto GPU + +- BCE_WEIGHTS -> the class weights for the Binary Cross Entropy loss + +## Training + +After configure config.py, you can start to train by running + +`python train.py` + +We also employ tensorboard to visualize the training process. + +`tensorboard --logdir=runs/`

