Downloads: 1
Switch to unified view
| a/README.md | b/README.md | ||
|---|---|---|---|
| 1 | --- |
1 | ---
|
| 2 | jupyter: |
2 | jupyter:
|
| 3 | jupytext: |
3 | jupytext:
|
| 4 | formats: ipynb,md |
4 | formats: ipynb,md
|
| 5 | text_representation: |
5 | text_representation:
|
| 6 | extension: .md |
6 | extension: .md
|
| 7 | format_name: markdown |
7 | format_name: markdown
|
| 8 | format_version: '1.3' |
8 | format_version: '1.3'
|
| 9 | jupytext_version: 1.14.4 |
9 | jupytext_version: 1.14.4
|
| 10 | kernelspec: |
10 | kernelspec:
|
| 11 | display_name: Python 3 (ipykernel) |
11 | display_name: Python 3 (ipykernel)
|
| 12 | language: python |
12 | language: python
|
| 13 | name: python3 |
13 | name: python3
|
| 14 | --- |
14 | --- |
| 15 | 15 | ||
| 16 | <!-- #region --> |
16 | <!-- #region -->
|
| 17 | # 3D Image Classification |
17 | # 3D Image Classification |
| 18 | 18 | ||
| 19 | Learn how to train a 3D convolutional neural network (3D CNN) to predict presence of pneumonia - based on [Tutorial on 3D Image Classification](https://keras.io/examples/vision/3D_image_classification/) by [Hasib Zunair](https://github.com/hasibzunair). |
19 | Learn how to train a 3D convolutional neural network (3D CNN) to predict presence of pneumonia - based on [Tutorial on 3D Image Classification](https://keras.io/examples/vision/3D_image_classification/) by [Hasib Zunair](https://github.com/hasibzunair). |
| 20 | 20 | ||
| 21 | 21 | ||
| 22 | > __Dataset__: [MosMedData: Chest CT Scans with COVID-19 Related Findings Dataset](https://www.medrxiv.org/content/10.1101/2020.05.20.20100362v1) :: This dataset contains anonymised human lung computed tomography (CT) scans with COVID-19 related findings, as well as without such findings. A small subset of studies has been annotated with binary pixel masks depicting regions of interests (ground-glass opacifications and consolidations). CT scans were obtained between 1st of March, 2020 and 25th of April, 2020, and provided by municipal hospitals in Moscow, Russia. |
22 | __Dataset__: [MosMedData: Chest CT Scans with COVID-19 Related Findings Dataset](https://www.medrxiv.org/content/10.1101/2020.05.20.20100362v1) :: This dataset contains anonymised human lung computed tomography (CT) scans with COVID-19 related findings, as well as without such findings. A small subset of studies has been annotated with binary pixel masks depicting regions of interests (ground-glass opacifications and consolidations). CT scans were obtained between 1st of March, 2020 and 25th of April, 2020, and provided by municipal hospitals in Moscow, Russia.
|
| 23 | <!-- #endregion --> |
23 | <!-- #endregion --> |
| 24 | 24 | ||
| 25 | ## Verify GPU Support |
25 | ## Verify GPU Support |
| 26 | 26 | ||
| 27 | ```python |
27 | ```python
|
| 28 | # importing tensorflow |
28 | # importing tensorflow
|
| 29 | import tensorflow as tf |
29 | import tensorflow as tf |
| 30 | 30 | ||
| 31 | device_name = tf.test.gpu_device_name() |
31 | device_name = tf.test.gpu_device_name()
|
| 32 | print('Active GPU :: {}'.format(device_name)) |
32 | print('Active GPU :: {}'.format(device_name))
|
| 33 | # Active GPU :: /device:GPU:0 |
33 | # Active GPU :: /device:GPU:0
|
| 34 | ``` |
34 | ``` |
| 35 | 35 | ||
| 36 | ## Import Dependencies |
36 | ## Import Dependencies |
| 37 | 37 | ||
| 38 | ```python |
38 | ```python
|
| 39 | import matplotlib.pyplot as plt |
39 | import matplotlib.pyplot as plt
|
| 40 | import nibabel as nib |
40 | import nibabel as nib
|
| 41 | import numpy as np |
41 | import numpy as np
|
| 42 | import os |
42 | import os
|
| 43 | import random |
43 | import random
|
| 44 | from scipy import ndimage |
44 | from scipy import ndimage
|
| 45 | from sklearn.model_selection import train_test_split |
45 | from sklearn.model_selection import train_test_split
|
| 46 | from tensorflow import keras |
46 | from tensorflow import keras
|
| 47 | from tensorflow.keras import layers |
47 | from tensorflow.keras import layers
|
| 48 | from tensorflow.keras.utils import plot_model |
48 | from tensorflow.keras.utils import plot_model
|
| 49 | ``` |
49 | ``` |
| 50 | 50 | ||
| 51 | ```python |
51 | ```python
|
| 52 | # helper functions |
52 | # helper functions
|
| 53 | from helper import (read_scan, |
53 | from helper import (read_scan,
|
| 54 | normalize, |
54 | normalize,
|
| 55 | resize_volume, |
55 | resize_volume,
|
| 56 | process_scan, |
56 | process_scan,
|
| 57 | rotate, |
57 | rotate,
|
| 58 | train_preprocessing, |
58 | train_preprocessing,
|
| 59 | validation_preprocessing, |
59 | validation_preprocessing,
|
| 60 | plot_slices, |
60 | plot_slices,
|
| 61 | build_model) |
61 | build_model)
|
| 62 | ``` |
62 | ``` |
| 63 | 63 | ||
| 64 | ## Import Dataset |
64 | ## Import Dataset |
| 65 | 65 | ||
| 66 | ```python |
66 | ```python
|
| 67 | # download from https://github.com/hasibzunair/3D-image-classification-tutorial/releases/ |
67 | # download from https://github.com/hasibzunair/3D-image-classification-tutorial/releases/
|
| 68 | data_dir = './dataset' |
68 | data_dir = './dataset'
|
| 69 | no_pneumonia = os.path.join(data_dir, 'no_viral_pneumonia') |
69 | no_pneumonia = os.path.join(data_dir, 'no_viral_pneumonia')
|
| 70 | with_pneumonia = os.path.join(data_dir, 'with_viral_pneumonia') |
70 | with_pneumonia = os.path.join(data_dir, 'with_viral_pneumonia') |
| 71 | 71 | ||
| 72 | normal_scan_paths = [ |
72 | normal_scan_paths = [
|
| 73 | os.path.join(no_pneumonia, i) |
73 | os.path.join(no_pneumonia, i)
|
| 74 | for i in os.listdir(no_pneumonia) |
74 | for i in os.listdir(no_pneumonia)
|
| 75 | ] |
75 | ]
|
| 76 | print('INFO :: CT Scans with normal lung tissue:', len(normal_scan_paths)) |
76 | print('INFO :: CT Scans with normal lung tissue:', len(normal_scan_paths)) |
| 77 | 77 | ||
| 78 | abnormal_scan_paths = [ |
78 | abnormal_scan_paths = [
|
| 79 | os.path.join(with_pneumonia, i) |
79 | os.path.join(with_pneumonia, i)
|
| 80 | for i in os.listdir(with_pneumonia) |
80 | for i in os.listdir(with_pneumonia)
|
| 81 | ] |
81 | ]
|
| 82 | print('INFO :: CT Scans with abnormal lung tissue:', len(abnormal_scan_paths)) |
82 | print('INFO :: CT Scans with abnormal lung tissue:', len(abnormal_scan_paths)) |
| 83 | 83 | ||
| 84 | # INFO :: CT Scans with normal lung tissue: 100 |
84 | # INFO :: CT Scans with normal lung tissue: 100
|
| 85 | # INFO :: CT Scans with abnormal lung tissue: 100 |
85 | # INFO :: CT Scans with abnormal lung tissue: 100
|
| 86 | ``` |
86 | ``` |
| 87 | 87 | ||
| 88 | ## Visualize Dataset |
88 | ## Visualize Dataset |
| 89 | 89 | ||
| 90 | ```python |
90 | ```python
|
| 91 | img_normal = nib.load(normal_scan_paths[0]) |
91 | img_normal = nib.load(normal_scan_paths[0])
|
| 92 | img_normal_array = img_normal.get_fdata() |
92 | img_normal_array = img_normal.get_fdata() |
| 93 | 93 | ||
| 94 | img_abnormal = nib.load(abnormal_scan_paths[0]) |
94 | img_abnormal = nib.load(abnormal_scan_paths[0])
|
| 95 | img_abnormal_array = img_abnormal.get_fdata() |
95 | img_abnormal_array = img_abnormal.get_fdata() |
| 96 | 96 | ||
| 97 | plt.figure(figsize=(30,10)) |
97 | plt.figure(figsize=(30,10)) |
| 98 | 98 | ||
| 99 | for i in range(6): |
99 | for i in range(6):
|
| 100 | plt.subplot(2, 6, i+1) |
100 | plt.subplot(2, 6, i+1)
|
| 101 | plt.imshow(img_normal_array[:, :, i], cmap='Blues') |
101 | plt.imshow(img_normal_array[:, :, i], cmap='Blues')
|
| 102 | plt.axis('off') |
102 | plt.axis('off')
|
| 103 | plt.title('Slice {} - Normal'.format(i)) |
103 | plt.title('Slice {} - Normal'.format(i))
|
| 104 | 104 |
|
|
| 105 | plt.subplot(2, 6, 6+i+1) |
105 | plt.subplot(2, 6, 6+i+1)
|
| 106 | plt.imshow(img_abnormal_array[:, :, i], cmap='Reds') |
106 | plt.imshow(img_abnormal_array[:, :, i], cmap='Reds')
|
| 107 | plt.axis('off') |
107 | plt.axis('off')
|
| 108 | plt.title('Slice {} - Abnormal'.format(i)) |
108 | plt.title('Slice {} - Abnormal'.format(i))
|
| 109 | ``` |
109 | ``` |
| 110 | 110 | ||
| 111 |  |
111 | 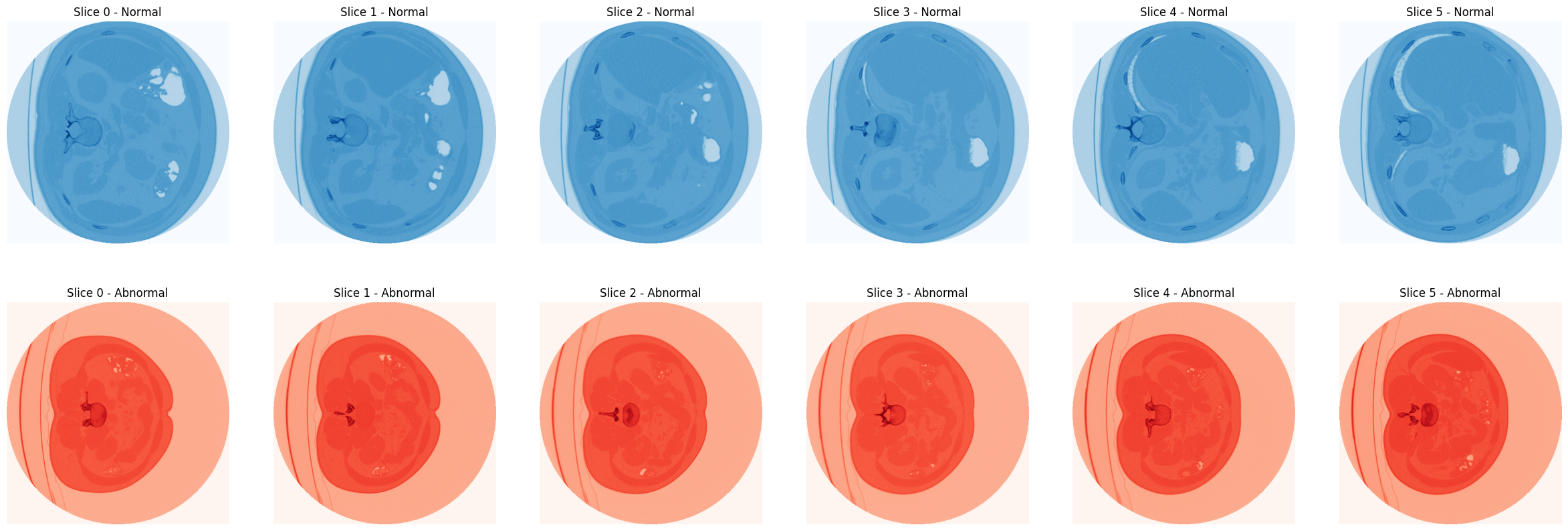 |
| 112 | 112 | ||
| 113 | 113 | ||
| 114 | ## Data Pre-processing |
114 | ## Data Pre-processing |
| 115 | 115 | ||
| 116 | ### Normalization |
116 | ### Normalization |
| 117 | 117 | ||
| 118 | ```python |
118 | ```python
|
| 119 | # Read and process the scans. |
119 | # Read and process the scans.
|
| 120 | # Each scan is resized across height, width, and depth and rescaled. |
120 | # Each scan is resized across height, width, and depth and rescaled.
|
| 121 | abnormal_scans = np.array([process_scan(path) for path in abnormal_scan_paths]) |
121 | abnormal_scans = np.array([process_scan(path) for path in abnormal_scan_paths])
|
| 122 | normal_scans = np.array([process_scan(path) for path in normal_scan_paths]) |
122 | normal_scans = np.array([process_scan(path) for path in normal_scan_paths])
|
| 123 | ``` |
123 | ``` |
| 124 | 124 | ||
| 125 | ```python |
125 | ```python
|
| 126 | # For the CT scans having presence of viral pneumonia |
126 | # For the CT scans having presence of viral pneumonia
|
| 127 | # assign 1, for the normal ones assign 0. |
127 | # assign 1, for the normal ones assign 0.
|
| 128 | abnormal_labels = np.array([1 for _ in range(len(abnormal_scans))]) |
128 | abnormal_labels = np.array([1 for _ in range(len(abnormal_scans))])
|
| 129 | normal_labels = np.array([0 for _ in range(len(normal_scans))]) |
129 | normal_labels = np.array([0 for _ in range(len(normal_scans))])
|
| 130 | ``` |
130 | ``` |
| 131 | 131 | ||
| 132 | ### Train Test Split |
132 | ### Train Test Split |
| 133 | 133 | ||
| 134 | ```python |
134 | ```python
|
| 135 | X = np.concatenate((abnormal_scans, normal_scans), axis=0) |
135 | X = np.concatenate((abnormal_scans, normal_scans), axis=0)
|
| 136 | Y = np.concatenate((abnormal_labels, normal_labels), axis=0) |
136 | Y = np.concatenate((abnormal_labels, normal_labels), axis=0) |
| 137 | 137 | ||
| 138 | x_train, x_val, y_train, y_val = train_test_split(X, Y, test_size=0.3, random_state=42) |
138 | x_train, x_val, y_train, y_val = train_test_split(X, Y, test_size=0.3, random_state=42)
|
| 139 | print('INFO :: Train / Test Samples - %d / %d' % (x_train.shape[0], x_val.shape[0])) |
139 | print('INFO :: Train / Test Samples - %d / %d' % (x_train.shape[0], x_val.shape[0]))
|
| 140 | # INFO :: Train / Test Samples - 140 / 60 |
140 | # INFO :: Train / Test Samples - 140 / 60
|
| 141 | ``` |
141 | ``` |
| 142 | 142 | ||
| 143 | ### Data Augmentation |
143 | ### Data Augmentation |
| 144 | 144 | ||
| 145 | 145 | ||
| 146 | #### Data Loader |
146 | #### Data Loader |
| 147 | 147 | ||
| 148 | ```python |
148 | ```python
|
| 149 | # Define data loaders. |
149 | # Define data loaders.
|
| 150 | train_loader = tf.data.Dataset.from_tensor_slices((x_train, y_train)) |
150 | train_loader = tf.data.Dataset.from_tensor_slices((x_train, y_train))
|
| 151 | validation_loader = tf.data.Dataset.from_tensor_slices((x_val, y_val)) |
151 | validation_loader = tf.data.Dataset.from_tensor_slices((x_val, y_val))
|
| 152 | batch_size = 2 |
152 | batch_size = 2 |
| 153 | 153 | ||
| 154 | # Augment the on the fly during training. |
154 | # Augment the on the fly during training.
|
| 155 | train_dataset = ( |
155 | train_dataset = (
|
| 156 | train_loader.shuffle(len(x_train)) |
156 | train_loader.shuffle(len(x_train))
|
| 157 | .map(train_preprocessing) |
157 | .map(train_preprocessing)
|
| 158 | .batch(batch_size) |
158 | .batch(batch_size)
|
| 159 | .prefetch(2) |
159 | .prefetch(2)
|
| 160 | ) |
160 | )
|
| 161 | # Only rescale. |
161 | # Only rescale.
|
| 162 | validation_dataset = ( |
162 | validation_dataset = (
|
| 163 | validation_loader.shuffle(len(x_val)) |
163 | validation_loader.shuffle(len(x_val))
|
| 164 | .map(validation_preprocessing) |
164 | .map(validation_preprocessing)
|
| 165 | .batch(batch_size) |
165 | .batch(batch_size)
|
| 166 | .prefetch(2) |
166 | .prefetch(2)
|
| 167 | ) |
167 | )
|
| 168 | ``` |
168 | ``` |
| 169 | 169 | ||
| 170 | ### Visualizing Augmented Datasets |
170 | ### Visualizing Augmented Datasets |
| 171 | 171 | ||
| 172 | ```python |
172 | ```python
|
| 173 | data = train_dataset.take(1) |
173 | data = train_dataset.take(1)
|
| 174 | images, labels = list(data)[0] |
174 | images, labels = list(data)[0]
|
| 175 | images = images.numpy() |
175 | images = images.numpy()
|
| 176 | image = images[0] |
176 | image = images[0]
|
| 177 | print("CT Scan Dims:", image.shape) |
177 | print("CT Scan Dims:", image.shape)
|
| 178 | # CT Scan Dims: (128, 128, 64, 1) |
178 | # CT Scan Dims: (128, 128, 64, 1)
|
| 179 | plt.imshow(np.squeeze(image[:, :, 30]), cmap="gray") |
179 | plt.imshow(np.squeeze(image[:, :, 30]), cmap="gray") |
| 180 | 180 | ||
| 181 | # Visualize montage of slices. |
181 | # Visualize montage of slices.
|
| 182 | # 4 rows and 10 columns for 100 slices of the CT scan. |
182 | # 4 rows and 10 columns for 100 slices of the CT scan.
|
| 183 | plot_slices(4, 10, 128, 128, image[:, :, :40]) |
183 | plot_slices(4, 10, 128, 128, image[:, :, :40])
|
| 184 | ``` |
184 | ``` |
| 185 | 185 | ||
| 186 |  |
186 |  |
| 187 | 187 | ||
| 188 |  |
188 | 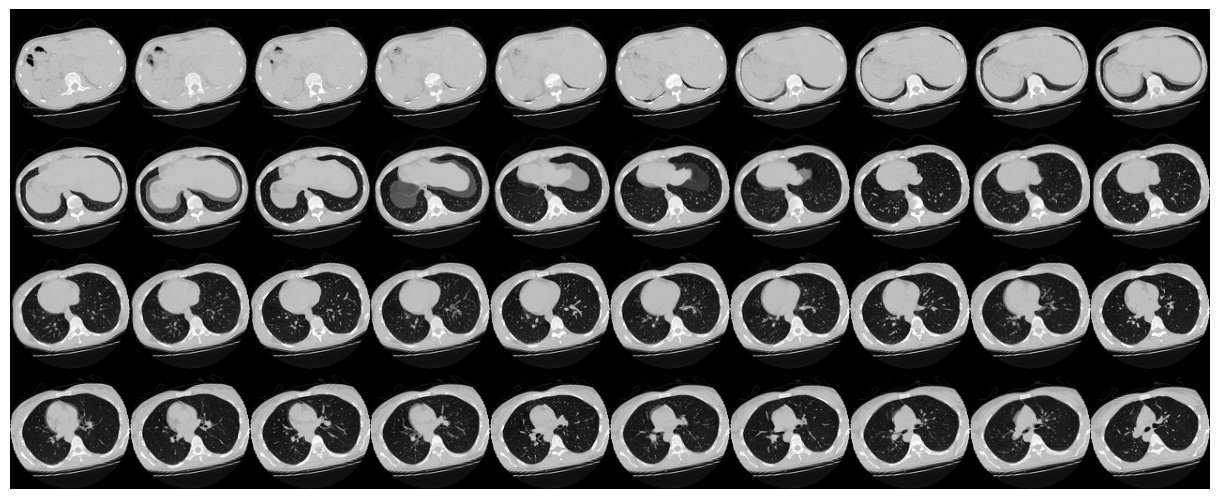 |
| 189 | 189 | ||
| 190 | 190 | ||
| 191 | ## Building the Model |
191 | ## Building the Model |
| 192 | 192 | ||
| 193 | ```python |
193 | ```python
|
| 194 | model = build_model(width=128, height=128, depth=64) |
194 | model = build_model(width=128, height=128, depth=64)
|
| 195 | model.summary() |
195 | model.summary()
|
| 196 | ``` |
196 | ``` |
| 197 | 197 | ||
| 198 | <!-- #region --> |
198 | <!-- #region -->
|
| 199 | ```bash |
199 | ```bash
|
| 200 | Model: "3dctscan" |
200 | Model: "3dctscan"
|
| 201 | _________________________________________________________________ |
201 | _________________________________________________________________
|
| 202 | Layer (type) Output Shape Param # |
202 | Layer (type) Output Shape Param #
|
| 203 | ================================================================= |
203 | =================================================================
|
| 204 | input_3 (InputLayer) [(None, 128, 128, 64, 1) 0 |
204 | input_3 (InputLayer) [(None, 128, 128, 64, 1) 0
|
| 205 | ] |
205 | ]
|
| 206 | 206 |
|
|
| 207 | conv3d_9 (Conv3D) (None, 126, 126, 62, 64) 1792 |
207 | conv3d_9 (Conv3D) (None, 126, 126, 62, 64) 1792
|
| 208 | 208 |
|
|
| 209 | max_pooling3d_8 (MaxPooling (None, 63, 63, 31, 64) 0 |
209 | max_pooling3d_8 (MaxPooling (None, 63, 63, 31, 64) 0
|
| 210 | 3D) |
210 | 3D)
|
| 211 | 211 |
|
|
| 212 | batch_normalization_8 (Batc (None, 63, 63, 31, 64) 256 |
212 | batch_normalization_8 (Batc (None, 63, 63, 31, 64) 256
|
| 213 | hNormalization) |
213 | hNormalization)
|
| 214 | 214 |
|
|
| 215 | conv3d_10 (Conv3D) (None, 61, 61, 29, 64) 110656 |
215 | conv3d_10 (Conv3D) (None, 61, 61, 29, 64) 110656
|
| 216 | 216 |
|
|
| 217 | max_pooling3d_9 (MaxPooling (None, 30, 30, 14, 64) 0 |
217 | max_pooling3d_9 (MaxPooling (None, 30, 30, 14, 64) 0
|
| 218 | 3D) |
218 | 3D)
|
| 219 | 219 |
|
|
| 220 | batch_normalization_9 (Batc (None, 30, 30, 14, 64) 256 |
220 | batch_normalization_9 (Batc (None, 30, 30, 14, 64) 256
|
| 221 | hNormalization) |
221 | hNormalization)
|
| 222 | 222 |
|
|
| 223 | conv3d_11 (Conv3D) (None, 28, 28, 12, 128) 221312 |
223 | conv3d_11 (Conv3D) (None, 28, 28, 12, 128) 221312
|
| 224 | 224 |
|
|
| 225 | max_pooling3d_10 (MaxPoolin (None, 14, 14, 6, 128) 0 |
225 | max_pooling3d_10 (MaxPoolin (None, 14, 14, 6, 128) 0
|
| 226 | g3D) |
226 | g3D)
|
| 227 | 227 |
|
|
| 228 | batch_normalization_10 (Bat (None, 14, 14, 6, 128) 512 |
228 | batch_normalization_10 (Bat (None, 14, 14, 6, 128) 512
|
| 229 | chNormalization) |
229 | chNormalization)
|
| 230 | 230 |
|
|
| 231 | conv3d_12 (Conv3D) (None, 12, 12, 4, 256) 884992 |
231 | conv3d_12 (Conv3D) (None, 12, 12, 4, 256) 884992
|
| 232 | 232 |
|
|
| 233 | max_pooling3d_11 (MaxPoolin (None, 6, 6, 2, 256) 0 |
233 | max_pooling3d_11 (MaxPoolin (None, 6, 6, 2, 256) 0
|
| 234 | g3D) |
234 | g3D)
|
| 235 | 235 |
|
|
| 236 | batch_normalization_11 (Bat (None, 6, 6, 2, 256) 1024 |
236 | batch_normalization_11 (Bat (None, 6, 6, 2, 256) 1024
|
| 237 | chNormalization) |
237 | chNormalization)
|
| 238 | 238 |
|
|
| 239 | global_average_pooling3d_1 (None, 256) 0 |
239 | global_average_pooling3d_1 (None, 256) 0
|
| 240 | (GlobalAveragePooling3D) |
240 | (GlobalAveragePooling3D)
|
| 241 | 241 |
|
|
| 242 | dense_2 (Dense) (None, 512) 131584 |
242 | dense_2 (Dense) (None, 512) 131584
|
| 243 | 243 |
|
|
| 244 | dropout_1 (Dropout) (None, 512) 0 |
244 | dropout_1 (Dropout) (None, 512) 0
|
| 245 | 245 |
|
|
| 246 | dense_3 (Dense) (None, 1) 513 |
246 | dense_3 (Dense) (None, 1) 513
|
| 247 | 247 |
|
|
| 248 | ================================================================= |
248 | =================================================================
|
| 249 | Total params: 1,352,897 |
249 | Total params: 1,352,897
|
| 250 | Trainable params: 1,351,873 |
250 | Trainable params: 1,351,873
|
| 251 | Non-trainable params: 1,024 |
251 | Non-trainable params: 1,024
|
| 252 | _________________________________________________________________ |
252 | _________________________________________________________________
|
| 253 | ``` |
253 | ```
|
| 254 | <!-- #endregion --> |
254 | <!-- #endregion --> |
| 255 | 255 | ||
| 256 | ### Compile the Model |
256 | ### Compile the Model |
| 257 | 257 | ||
| 258 | ```python |
258 | ```python
|
| 259 | initial_learning_rate = 0.0001 |
259 | initial_learning_rate = 0.0001 |
| 260 | 260 | ||
| 261 | lr_schedule = keras.optimizers.schedules.ExponentialDecay( |
261 | lr_schedule = keras.optimizers.schedules.ExponentialDecay(
|
| 262 | initial_learning_rate, |
262 | initial_learning_rate,
|
| 263 | decay_steps=100000, |
263 | decay_steps=100000,
|
| 264 | decay_rate=0.96, |
264 | decay_rate=0.96,
|
| 265 | staircase=True) |
265 | staircase=True) |
| 266 | 266 | ||
| 267 | model.compile( |
267 | model.compile(
|
| 268 | loss='binary_crossentropy', |
268 | loss='binary_crossentropy',
|
| 269 | optimizer=keras.optimizers.Adam(learning_rate=lr_schedule), |
269 | optimizer=keras.optimizers.Adam(learning_rate=lr_schedule),
|
| 270 | metrics=['acc'] |
270 | metrics=['acc']
|
| 271 | ) |
271 | )
|
| 272 | ``` |
272 | ``` |
| 273 | 273 | ||
| 274 | ### Callbacks |
274 | ### Callbacks |
| 275 | 275 | ||
| 276 | ```python |
276 | ```python
|
| 277 | cp_cb = keras.callbacks.ModelCheckpoint( |
277 | cp_cb = keras.callbacks.ModelCheckpoint(
|
| 278 | './checkpoints/3dct_weights.h5', |
278 | './checkpoints/3dct_weights.h5',
|
| 279 | save_best_only=True |
279 | save_best_only=True
|
| 280 | ) |
280 | ) |
| 281 | 281 | ||
| 282 | es_cb = keras.callbacks.EarlyStopping( |
282 | es_cb = keras.callbacks.EarlyStopping(
|
| 283 | monitor='val_acc', |
283 | monitor='val_acc',
|
| 284 | patience=15 |
284 | patience=15
|
| 285 | ) |
285 | )
|
| 286 | ``` |
286 | ``` |
| 287 | 287 | ||
| 288 | ## Model Training |
288 | ## Model Training |
| 289 | 289 | ||
| 290 | ```python |
290 | ```python
|
| 291 | epochs = 100 |
291 | epochs = 100 |
| 292 | 292 | ||
| 293 | model.fit( |
293 | model.fit(
|
| 294 | train_dataset, |
294 | train_dataset,
|
| 295 | validation_data=validation_dataset, |
295 | validation_data=validation_dataset,
|
| 296 | epochs=epochs, |
296 | epochs=epochs,
|
| 297 | shuffle=True, |
297 | shuffle=True,
|
| 298 | verbose=2, |
298 | verbose=2,
|
| 299 | callbacks=[cp_cb, es_cb] |
299 | callbacks=[cp_cb, es_cb]
|
| 300 | ) |
300 | )
|
| 301 | # Epoch 46/100 |
301 | # Epoch 46/100
|
| 302 | # 70/70 - 22s - loss: 0.3383 - acc: 0.8429 - val_loss: 0.8225 - val_acc: 0.6833 - 22s/epoch - 313ms/step |
302 | # 70/70 - 22s - loss: 0.3383 - acc: 0.8429 - val_loss: 0.8225 - val_acc: 0.6833 - 22s/epoch - 313ms/step
|
| 303 | ``` |
303 | ``` |
| 304 | 304 | ||
| 305 | ### Visualizing Model Performance |
305 | ### Visualizing Model Performance |
| 306 | 306 | ||
| 307 | ```python |
307 | ```python
|
| 308 | fig, ax = plt.subplots(1, 2, figsize=(20, 3)) |
308 | fig, ax = plt.subplots(1, 2, figsize=(20, 3))
|
| 309 | ax = ax.ravel() |
309 | ax = ax.ravel() |
| 310 | 310 | ||
| 311 | for i, metric in enumerate(["acc", "loss"]): |
311 | for i, metric in enumerate(["acc", "loss"]):
|
| 312 | ax[i].plot(model.history.history[metric]) |
312 | ax[i].plot(model.history.history[metric])
|
| 313 | ax[i].plot(model.history.history["val_" + metric]) |
313 | ax[i].plot(model.history.history["val_" + metric])
|
| 314 | ax[i].set_title("Model {}".format(metric)) |
314 | ax[i].set_title("Model {}".format(metric))
|
| 315 | ax[i].set_xlabel("epochs") |
315 | ax[i].set_xlabel("epochs")
|
| 316 | ax[i].set_ylabel(metric) |
316 | ax[i].set_ylabel(metric)
|
| 317 | ax[i].legend(["train", "val"]) |
317 | ax[i].legend(["train", "val"])
|
| 318 | ``` |
318 | ``` |
| 319 | 319 | ||
| 320 |  |
320 | 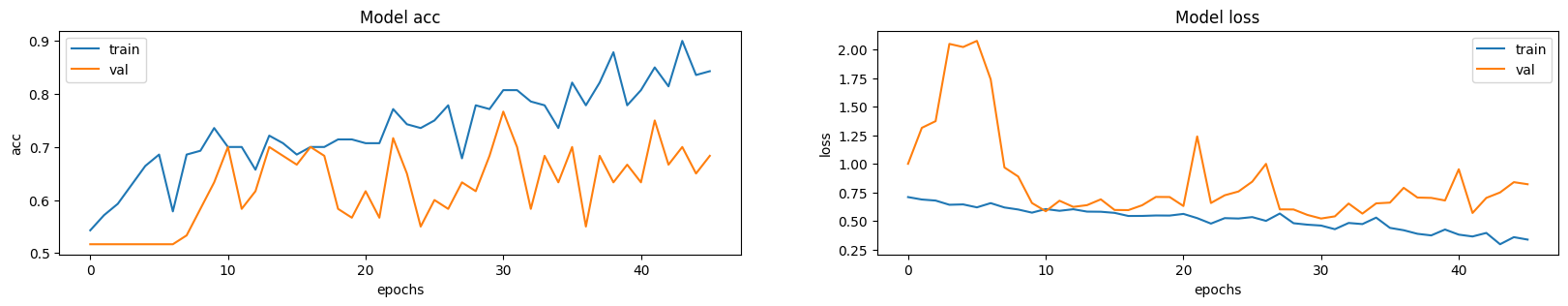 |
| 321 | 321 | ||
| 322 | 322 | ||
| 323 | ## Loading Best Training Weights |
323 | ## Loading Best Training Weights |
| 324 | 324 | ||
| 325 | ```python |
325 | ```python
|
| 326 | model.load_weights('./checkpoints/3dct_weights.h5') |
326 | model.load_weights('./checkpoints/3dct_weights.h5')
|
| 327 | ``` |
327 | ``` |
| 328 | 328 | ||
| 329 | ## Make Predictions |
329 | ## Make Predictions |
| 330 | 330 | ||
| 331 | ```python |
331 | ```python
|
| 332 | pred_dataset = './predictions' |
332 | pred_dataset = './predictions'
|
| 333 | pred_paths = [os.path.join(pred_dataset, i) for i in os.listdir(pred_dataset)] |
333 | pred_paths = [os.path.join(pred_dataset, i) for i in os.listdir(pred_dataset)] |
| 334 | 334 | ||
| 335 | z_val = np.array([process_scan(path) for path in pred_paths]) |
335 | z_val = np.array([process_scan(path) for path in pred_paths]) |
| 336 | 336 | ||
| 337 | for i in range(len(z_val)): |
337 | for i in range(len(z_val)):
|
| 338 | prediction = model.predict(np.expand_dims(z_val[i], axis=0))[0] |
338 | prediction = model.predict(np.expand_dims(z_val[i], axis=0))[0]
|
| 339 | scores = [1 - prediction[0], prediction[0]] |
339 | scores = [1 - prediction[0], prediction[0]]
|
| 340 | class_names = ['normal', 'abnormal'] |
340 | class_names = ['normal', 'abnormal']
|
| 341 | 341 |
|
|
| 342 | pred_image = nib.load(pred_paths[i]) |
342 | pred_image = nib.load(pred_paths[i])
|
| 343 | pred_image_data = pred_image.get_fdata() |
343 | pred_image_data = pred_image.get_fdata() |
| 344 | 344 | ||
| 345 | normal_class = class_names[0], round(100*scores[0], 2) |
345 | normal_class = class_names[0], round(100*scores[0], 2)
|
| 346 | abnormal_class = class_names[1], round(100*scores[1], 2) |
346 | abnormal_class = class_names[1], round(100*scores[1], 2)
|
| 347 | annotation = normal_class + abnormal_class |
347 | annotation = normal_class + abnormal_class |
| 348 | 348 | ||
| 349 | plt.imshow(pred_image_data[:,:, pred_image_data.shape[2]//2], cmap='gray') |
349 | plt.imshow(pred_image_data[:,:, pred_image_data.shape[2]//2], cmap='gray')
|
| 350 | plt.title(os.path.basename(pred_paths[i])) |
350 | plt.title(os.path.basename(pred_paths[i]))
|
| 351 | plt.xlabel(annotation) |
351 | plt.xlabel(annotation)
|
| 352 | plt.show() |
352 | plt.show()
|
| 353 | ``` |
353 | ``` |
| 354 | 354 | ||
| 355 |  |
355 | 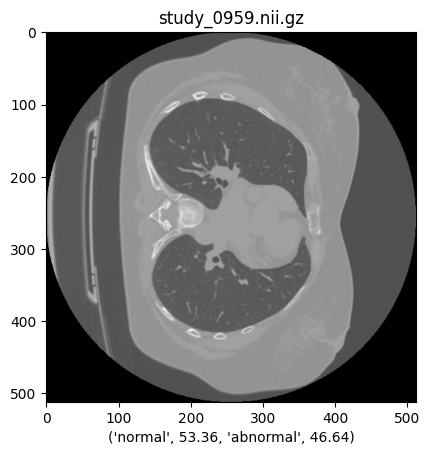 |
| 356 | 356 | ||
| 357 | ```python |
357 | ```python
|
| 358 | print(scores, class_names) |
358 | print(scores, class_names)
|
| 359 | ``` |
359 | ``` |
| 360 | 360 | ||
| 361 | ```python |
361 | ```python |
| 362 | 362 | ||
| 363 | ``` |
363 | ```
|

 Datasets
Datasets
 Models
Models