Downloads: 1
Diff of
/README.md
[000000]
..
[eac570]
Switch to side-by-side view
--- a +++ b/README.md @@ -0,0 +1,172 @@ +# 3D Neural Network for Lung Cancer Risk Prediction on CT Volumes + +<!--- +[](https://doi.org/10.5281/zenodo.3950478) +--> + +### Overview + +This repository contains my implementation of the "full-volume" model from the paper: + +[End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography.](https://doi.org/10.1038/s41591-019-0447-x)<br/> Ardila, D., Kiraly, A.P., Bharadwaj, S. et al. Nat Med 25, 954–961 (2019). + +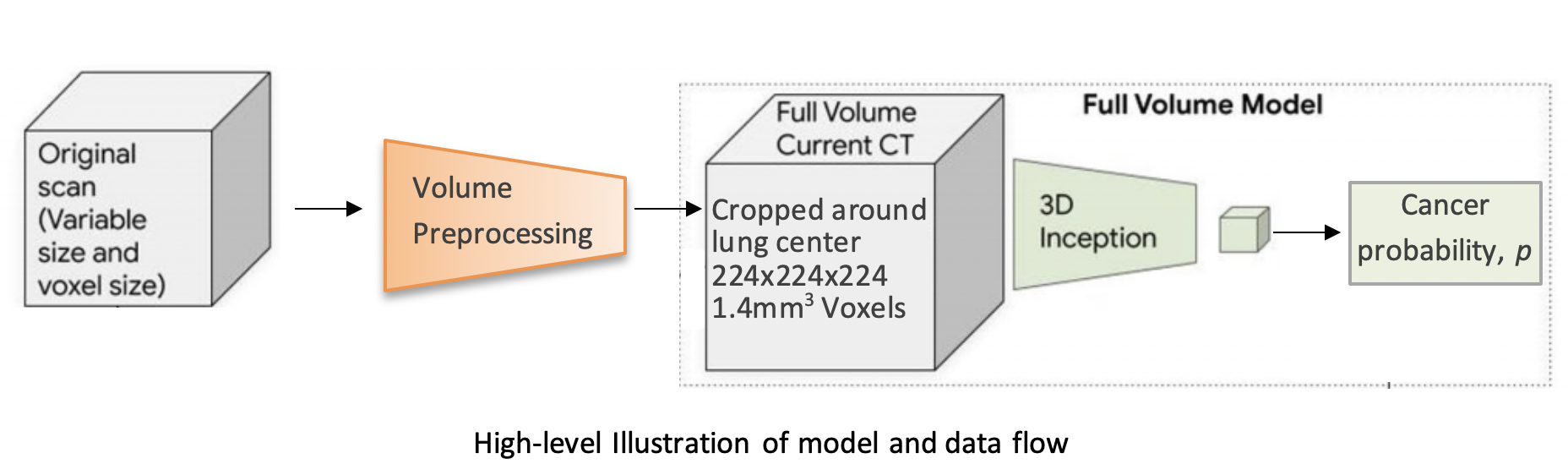 + +The model uses a three-dimensional (3D) CNN to perform end-to-end analysis of whole-CT volumes, using LDCT +volumes with pathology-confirmed cancer as training data. +The CNN architecture is an Inflated 3D ConvNet (I3D) ([Carreira and +Zisserman](http://openaccess.thecvf.com/content_cvpr_2017/html/Carreira_Quo_Vadis_Action_CVPR_2017_paper.html)). + +### High-level Presentation + +[](http://www.youtube.com/watch?v=KPBEpc_yDRk) + +### Detailed Report +For more detailed information, see this [report](https://arxiv.org/abs/2007.12898) on arXiv. + +### Data + +We use the NLST dataset which contains chest LDCT volumes with pathology-confirmed cancer evaluations. For description and access to the dataset refer to the [NCI website](https://biometry.nci.nih.gov/cdas/learn/nlst/images/). + +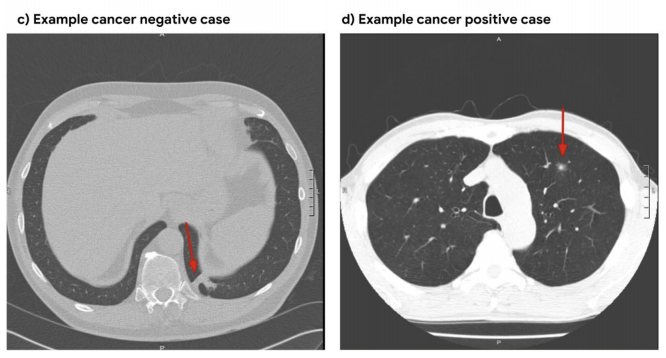 + +### Setup + +```bash +pip install lungs +``` + +### Running the code + +#### Inference + +```python +import lungs +lungs.predict('path/to/data') +``` + +From command line (with preprocessed data): + +```bash +python main.py --preprocessed --input path/to/preprocessed/data +``` + +#### Training + +The inputs to the training procedure are training and validation `.list` files containing coupled data - a volume path and its label in each row. +These `.list` files need to be generated beforehand using `preprocess.py`, as described in the next section. + +```python +import lungs +# train with default hyperparameters +lungs.train(train='path/to/train.list', val='path/to/val.list') +``` + +The `main.py` module contains training (fine-tuning) and inference procedures. +The inputs are preprocessed CT volumes, as produced by `preprocess.py`. +For usage example, refer to the arguments' description and default values in the bottom of `main.py`. + +### Data Preprocessing + +#### Preprocess volumes + +Each CT volume in NLST is a **directory** of DICOM files (each `.dcm` file is one slice of the CT). +The `preprocess.py` module accepts a directory `path/to/data` containing **multiple** such directories (volumes). +It performs several preprocessing steps, and writes each preprocessed volume as an `.npz` file in `path/to/data_preprocssed`. +These steps are based on [this](https://www.kaggle.com/gzuidhof/full-preprocessing-tutorial/notebook) tutorial, and include: + +- Resampling to a 1.5mm voxel size (slow) +- Coarse lung segmentation – used to compute lung center for alignment and reduction of problem space + +To save storage space, the following preprocessing steps are performed online (during training/inference): + +- Windowing – clip pixel values to focus on lung volume +- RGB normalization + +Example usage: + +```python +from lungs import preprocess +# Step 1: Preprocess all volumes, will save them to '/path/to/dataset_preprocessed' +preprocess.preprocess_all('/path/to/dataset') +``` + +#### Create train/val `.list` files + +Once the preprocessed data is ready, the next step is to split it randomly into train/val sets, +and save each set as a `.list` file of paths/labels, required for the training procedure. + +Example usage: + +```python +from lungs import preprocess +# Step 2: Split preprocessed data into train/val coupled `.list` files +preprocess.split(positives='/path/to/dataset_preprocessed/positives', + negatives='/path/to/dataset_preprocessed/negatives', + lists_dir='/path/to/write/lists', + split_ratio=0.7) +``` + +### Provided checkpoint + +By default, if the `ckpt` argument is not given, the model is initialized using our best fine-tuned checkpoint. +Due to limited storage and compute time, our checkpoint was trained on a small subset of NLST containing 1,045 volumes (34% positive). + +**Note that in order to obtain a general purpose prediction model, one would have to train on the full NLST dataset. Steps include:** + +- Gaining access to the [NLST dataset](https://biometry.nci.nih.gov/cdas/learn/nlst/images/) +- Downloading<sup>1</sup> ~6TB of positive and negative volumes (requires storage and a few days for downloading) +- Preprocessing (see Data Preprocessing section above) +- Training (requires a capable GPU) + +Even though we used a small subset of NLST, we still achieved a state-of-the-art AUC score of 0.892 on a validation set of 448 volumes. +This is comparable to the original paper's AUC for the full-volume model (see the paper's supplementary material), trained on 47,974 volumes (1.34% positive). + +To train this model we first initialized by bootstrapping the filters from the [ImageNet pre-trained 2D Inception-v1 model]((http://download.tensorflow.org/models/inception_v1_2016_08_28.tar.gz)) into 3D, as described in the I3D paper. +It was then fine-tuned on the preprocessed CT volumes to predict cancer within 1 year (binary classification). Each of these volumes was a large region cropped around the center of the bounding box, as determined by lung segmentation in the preprocessing step. + +For the training setup, we set the dropout keep_prob to 0.7, and trained in mini-batches of size of 2 (due to limited GPU memory). We used `tf.train.AdamOptimizer` with a small learning rate of 5e-5, (due to the small batch size) and stopped the training before overfitting started around epoch 37. +The focal loss function from the paper is provided in the code, but we did not experience improved results using it, compared to cross-entropy loss which was used instead. The likely reason is that our dataset was more balanced than the original paper's. + +The follwoing plots show loss, AUC, and accuracy progression during training, along with ROC curves for selected epochs: + +<img src="https://raw.githubusercontent.com/danielkorat/Lung-Cancer-Risk-Prediction/master/figures/loss.png" width="786" height="420"> +<img src="https://raw.githubusercontent.com/danielkorat/Lung-Cancer-Risk-Prediction/master/figures/auc_and_accuracy.png" width="786" height="420"> + +<img src="https://raw.githubusercontent.com/danielkorat/Lung-Cancer-Risk-Prediction/master/figures/epoch_10.png" width="270" height="270"><img src="https://raw.githubusercontent.com/danielkorat/Lung-Cancer-Risk-Prediction/master/figures/epoch_20.png" width="270" height="270"><img src="https://raw.githubusercontent.com/danielkorat/Lung-Cancer-Risk-Prediction/master/figures/epoch_32.png" width="270" height="270"> + +### Citation + +```bibtex +@software{korat-lung-cancer-pred, + author = {Daniel Korat}, + title = {{3D Neural Network for Lung Cancer Risk Prediction on CT Volumes}}, + month = jul, + year = 2020, + publisher = {Zenodo}, + version = {v1.0}, + doi = {10.5281/zenodo.3950478}, + url = {https://doi.org/10.5281/zenodo.3950478} +} +``` + +### Acknowledgments + +The author thanks the National Cancer Institute for access to NCI's data collected by the National Screening Trial (NLST). +The statements contained herein are solely those of the author and do not represent or imply concurrence or endorsement by NCI. + +<sup>1</sup> Downloading volumes is done by querying the TCIA website (instruction on NCI website). We used the following query filters: + + - `LungCancerDiagnosis.conflc == "Confirmed.."` (positive) or `"Confirmed Not..."` (negative) + - `SCTImageInfo.numberimages >= 130` (minimum number of slices) + - `SCTImageInfo.reconthickness < 5.0` (maximum slice thickness) + - `ScreeningResults.study_yr = x` (study year of volume, a number between 0 and 7) + - `LungCancerDiagnosis.cancyr = x` or `x + 1` (for positives: study year of patient's cancer diagnosis is equal to `study_yr` or 1 year later) + +### All Links + +[GitHub Pages](https://danielkorat.github.io/Lung-Cancer-Risk-Prediction) +[arXiv Report](https://arxiv.org/abs/2007.12898) +[Presentation Video](http://www.youtube.com/watch?v=KPBEpc_yDRk) +[Presentation PDF](https://drive.google.com/file/d/1Wya9drNmXwdwWiwzeoKxgHmQ3r19Xa6Q/view?usp=sharing) +DOI: [https://doi.org/10.5281/zenodo.3950478](https://doi.org/10.5281/zenodo.3950478)

 Datasets
Datasets
 Models
Models