Downloads: 1
Diff of
/README.md
[000000]
..
[a8f942]
Switch to unified view
| a | b/README.md | ||
|---|---|---|---|
| 1 | # Predicting age from the electrocardiogram and its usage as a mortality predictor |
||
| 2 | |||
| 3 | Scripts and modules for training and testing deep neural networks for ECG automatic classification. |
||
| 4 | Companion code to the paper "Deep neural network-estimated electrocardiographic age as a mortality predictor". |
||
| 5 | https://www.nature.com/articles/s41467-021-25351-7. |
||
| 6 | |||
| 7 | Citation: |
||
| 8 | ``` |
||
| 9 | Lima, E.M., Ribeiro, A.H., Paixão, G.M.M. et al. Deep neural network-estimated electrocardiographic age as a |
||
| 10 | mortality predictor. Nat Commun 12, 5117 (2021). https://doi.org/10.1038/s41467-021-25351-7. |
||
| 11 | ``` |
||
| 12 | |||
| 13 | Bibtex: |
||
| 14 | ```bibtex |
||
| 15 | @article{lima_deep_2021, |
||
| 16 | title = {Deep Neural Network Estimated Electrocardiographic-Age as a Mortality Predictor}, |
||
| 17 | author = {Lima, Emilly M. and Ribeiro, Ant{\^o}nio H. and Paix{\~a}o, Gabriela MM and Ribeiro, Manoel Horta and Filho, Marcelo M. Pinto and Gomes, Paulo R. and Oliveira, Derick M. and Sabino, Ester C. and Duncan, Bruce B. and Giatti, Luana and Barreto, Sandhi M. and Meira, Wagner and Sch{\"o}n, Thomas B. and Ribeiro, Antonio Luiz P.}, |
||
| 18 | year = {2021}, |
||
| 19 | journal = {Nature Communications}, |
||
| 20 | volume = {12}, |
||
| 21 | doi = {10.1038/s41467-021-25351-7}, |
||
| 22 | annotation = {medRxiv doi: 10.1101/2021.02.19.21251232},} |
||
| 23 | } |
||
| 24 | ``` |
||
| 25 | **OBS:** *The three first authors: Emilly M. Lima, Antônio H. Ribeiro, Gabriela M. M. Paixão contributed equally.* |
||
| 26 | |||
| 27 | |||
| 28 | |||
| 29 | # Data |
||
| 30 | |||
| 31 | Three different cohorts are used in the study: |
||
| 32 | |||
| 33 | 1. The `CODE` study cohort, with n=1,558,415 patients was used for training and testing: |
||
| 34 | - exams from 15% of the patients in this cohort were used for testing. This sub-cohort is refered as `CODE-15%`. |
||
| 35 | The `CODE-15\%` dataset is openly available: [doi: 10.5281/zenodo.4916206 ](https://doi.org/10.5281/zenodo.4916206). |
||
| 36 | - the remainign 85% of the patients were used for developing the neural network model. |
||
| 37 | The full CODE dataset that was used for training is available upon |
||
| 38 | request for research purposes: [doi: 10.17044/scilifelab.15169716](https://doi.org/10.17044/scilifelab.15169716) |
||
| 39 | 2. The `SaMi-Trop` cohort, with n=1,631 patients, is used for external validation. |
||
| 40 | - The dataset is openly available: [doi: 10.5281/zenodo.4905618](https://doi.org/10.5281/zenodo.4905618) |
||
| 41 | 3. The `ELSA-Brasil` cohort with n=14,236 patients, is also used for external validation. |
||
| 42 | - Request to the ELSA-Brasil cohort should be forward to the ELSA-Brasil Steering Committee. |
||
| 43 | |||
| 44 | # Training and evaluation |
||
| 45 | |||
| 46 | The code training and evaluation is implemented in Python, contains |
||
| 47 | the code for training and evaluating the age prediction model. |
||
| 48 | |||
| 49 | ## Model |
||
| 50 | |||
| 51 | The model used in the paper is a residual neural network. The architecture implementation |
||
| 52 | in pytorch is available in `resnet.py`. It follows closely |
||
| 53 | [this architecture](https://www.nature.com/articles/s41467-020-15432-4), except that there is no sigmoid at the last layer. |
||
| 54 | |||
| 55 | 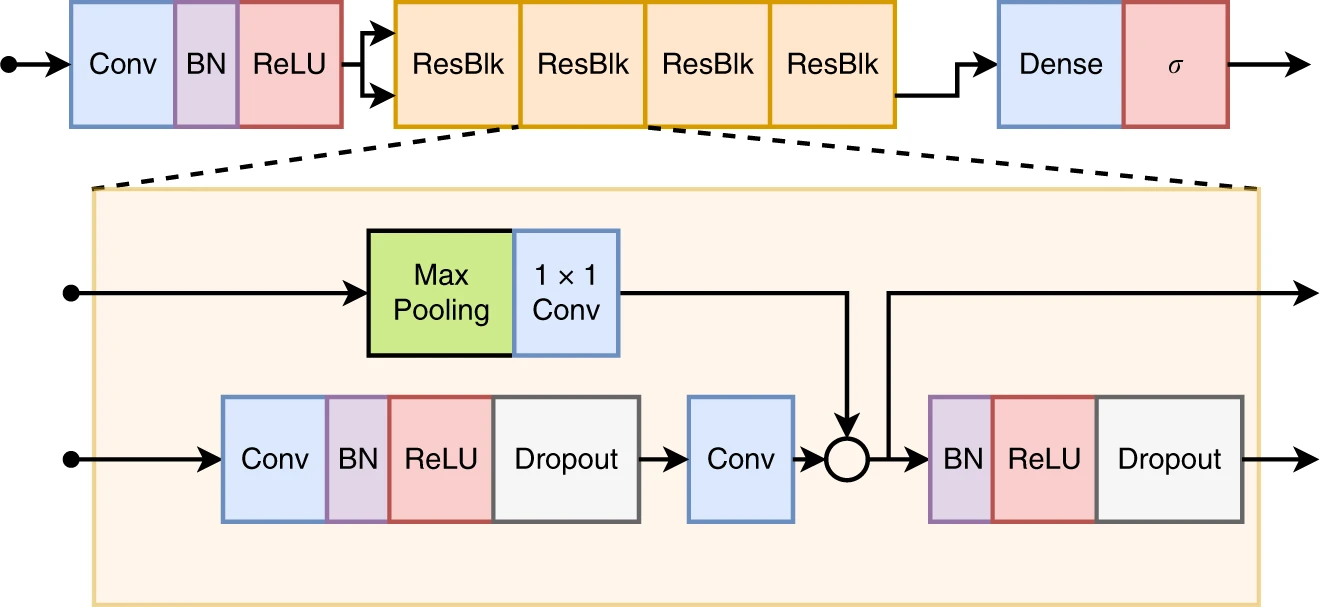 |
||
| 56 | |||
| 57 | The model can be trained using the script `train.py`. Alternatively, |
||
| 58 | pre-trained weighs trained on the code dataset for the model described in the paper |
||
| 59 | is available in [doi.org/10.5281/zenodo.4892365](https://doi.org/10.5281/zenodo.4892365) |
||
| 60 | in the following dropbox mirror |
||
| 61 | [here](https://www.dropbox.com/s/thvqwaryeo8uemo/model.zip?dl=0). |
||
| 62 | Using the command line, the weights can be downloaded using: |
||
| 63 | ``` |
||
| 64 | wget https://www.dropbox.com/s/thvqwaryeo8uemo/model.zip?dl=0 -O model.zip |
||
| 65 | unzip model.zip |
||
| 66 | ``` |
||
| 67 | - model input: `shape = (N, 12, 4096)`. The input tensor should contain the 4096 points of the ECG tracings sampled at 400Hz (i.e., a signal of approximately 10 seconds). Both in the training and in the test set, when the signal was not long enough, we filled the signal with zeros, so 4096 points were attained. The last dimension of the tensor contains points of the 12 different leads. The leads are ordered in the following order: {DI, DII, DIII, AVR, AVL, AVF, V1, V2, V3, V4, V5, V6}. All signal are represented as 32 bits floating point numbers at the scale 1e-4V: so if the signal is in V it should be multiplied by 1000 before feeding it to the neural network model. |
||
| 68 | - model output: `shape = (N, 1) `. With the entry being the predicted age from the ECG. |
||
| 69 | |||
| 70 | ## Requirements |
||
| 71 | |||
| 72 | This code was tested on Python 3 with Pytorch 1.2. It uses `numpy`, `pandas`, |
||
| 73 | `h5py` for loading and processing the data and `matplotlib` and `seaborn` |
||
| 74 | for the plots. See `requirements.txt` to see a full list of requirements |
||
| 75 | and library versions. |
||
| 76 | |||
| 77 | **For tensorflow users:** If you are interested in a tensorflow implementation, take a look in the repository: |
||
| 78 | https://github.com/antonior92/automatic-ecg-diagnosis. There we provide a tensorflow/keras implementation of the same |
||
| 79 | resnet-based model. The problem there is the abnormality classification from the ECG, nonetheless simple modifications |
||
| 80 | should suffice for dealing with age prediction |
||
| 81 | |||
| 82 | ## Folder content |
||
| 83 | |||
| 84 | |||
| 85 | - ``train.py``: Script for training the neural network. To train the neural network run: |
||
| 86 | ```bash |
||
| 87 | $ python train.py PATH_TO_HDF5 PATH_TO_CSV |
||
| 88 | ``` |
||
| 89 | |||
| 90 | |||
| 91 | - ``evaluate.py``: Script for generating the neural network predictions on a given dataset. |
||
| 92 | ```bash |
||
| 93 | $ python evaluate.py PATH_TO_MODEL PATH_TO_HDF5_ECG_TRACINGS --output PATH_TO_OUTPUT_FILE |
||
| 94 | ``` |
||
| 95 | |||
| 96 | |||
| 97 | - ``resnet.py``: Auxiliary module that defines the architecture of the deep neural network. |
||
| 98 | |||
| 99 | |||
| 100 | - ``formulate_problem.py``: Script that separate patients into training, validation and |
||
| 101 | ```bash |
||
| 102 | $ python predict.py PATH_TO_CSV |
||
| 103 | ``` |
||
| 104 | |||
| 105 | - ``plot_learning_curves.py``: Auxiliary script that plots learning curve of the model. |
||
| 106 | ```bash |
||
| 107 | $ python plot_learning_curves.py PATH_TO_MODEL/history.csv |
||
| 108 | ``` |
||
| 109 | |||
| 110 | OBS: Some scripts depend on the `resnet.py` and `dataloader.py` modules. So we recomend |
||
| 111 | the user to, either, run the scripts from within this folder or add it to your python path. |

 Datasets
Datasets
 Models
Models