Downloads: 1
Diff of
/README.md
[000000]
..
[c1a411]
Switch to unified view
| a | b/README.md | ||
|---|---|---|---|
| 1 | # Automatic ECG diagnosis using a deep neural network |
||
| 2 | Scripts and modules for training and testing deep neural networks for ECG automatic classification. |
||
| 3 | Companion code to the paper "Automatic diagnosis of the 12-lead ECG using a deep neural network". |
||
| 4 | https://www.nature.com/articles/s41467-020-15432-4. |
||
| 5 | |||
| 6 | -------- |
||
| 7 | |||
| 8 | Citation: |
||
| 9 | ``` |
||
| 10 | Ribeiro, A.H., Ribeiro, M.H., Paixão, G.M.M. et al. Automatic diagnosis of the 12-lead ECG using a deep neural network. |
||
| 11 | Nat Commun 11, 1760 (2020). https://doi.org/10.1038/s41467-020-15432-4 |
||
| 12 | ``` |
||
| 13 | |||
| 14 | Bibtex: |
||
| 15 | ``` |
||
| 16 | @article{ribeiro_automatic_2020, |
||
| 17 | title = {Automatic Diagnosis of the 12-Lead {{ECG}} Using a Deep Neural Network}, |
||
| 18 | author = {Ribeiro, Ant{\^o}nio H. and Ribeiro, Manoel Horta and Paix{\~a}o, Gabriela M. M. and Oliveira, Derick M. and Gomes, Paulo R. and Canazart, J{\'e}ssica A. and Ferreira, Milton P. S. and Andersson, Carl R. and Macfarlane, Peter W. and Meira Jr., Wagner and Sch{\"o}n, Thomas B. and Ribeiro, Antonio Luiz P.}, |
||
| 19 | year = {2020}, |
||
| 20 | volume = {11}, |
||
| 21 | pages = {1760}, |
||
| 22 | doi = {https://doi.org/10.1038/s41467-020-15432-4}, |
||
| 23 | journal = {Nature Communications}, |
||
| 24 | number = {1} |
||
| 25 | } |
||
| 26 | ``` |
||
| 27 | ----- |
||
| 28 | |||
| 29 | ## Requirements |
||
| 30 | |||
| 31 | This code was tested on Python 3 with Tensorflow `2.2`. There is an older branch ([`tensorflow-v1`](https://github.com/antonior92/automatic-ecg-diagnosis/tree/tensorflow-v1)) that |
||
| 32 | contain the code implementation for Tensorflow `1.15`. |
||
| 33 | |||
| 34 | **For pytorch users:** If you are interested in a pytorch implementation, take a look in the repository: https://github.com/antonior92/ecg-age-prediction. |
||
| 35 | There we provide a implementation in PyTorch of the same resnet-based model. The problem there is the age prediction from the ECG, nontheless simple modifications should suffice for dealing with abnormality classification. |
||
| 36 | |||
| 37 | ## Model |
||
| 38 | |||
| 39 | The model used in the paper is a residual neural. The neural network architecture implementation in Keras is available in ``model.py``. To print a summary of the model layers run: |
||
| 40 | ```bash |
||
| 41 | $ python model.py |
||
| 42 | ``` |
||
| 43 | |||
| 44 | 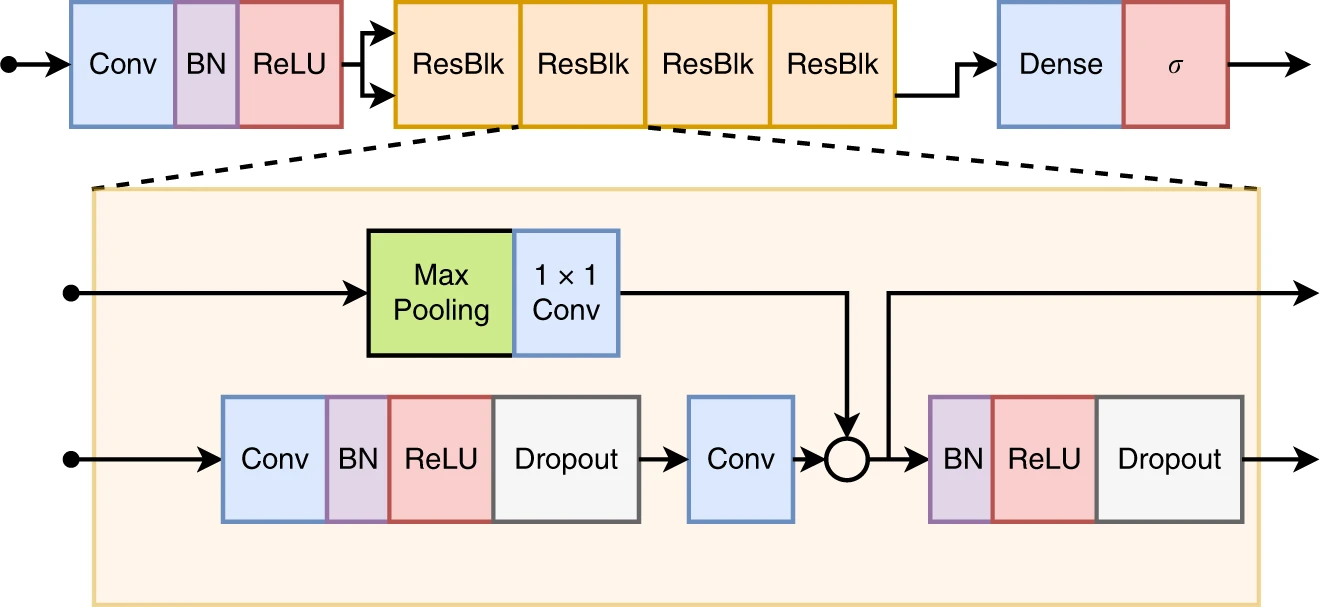 |
||
| 45 | |||
| 46 | The model receives an input tensor with dimension `(N, 4096, 12)`, and returns an output tensor with dimension `(N, 6)`, |
||
| 47 | for which `N` is the batch size. |
||
| 48 | |||
| 49 | The model can be trained using the script `train.py`. Alternatively, |
||
| 50 | pre-trained weighs for the models described in the paper are also |
||
| 51 | available in: https://doi.org/10.5281/zenodo.3625017. Or in the mirror dropbox |
||
| 52 | link [here](https://www.dropbox.com/s/5ar6j8u9v9a0rmh/model.zip?dl=0). |
||
| 53 | Using the command line, the weights can be downloaded using |
||
| 54 | ``` |
||
| 55 | wget https://www.dropbox.com/s/5ar6j8u9v9a0rmh/model.zip?dl=0 -O model.zip |
||
| 56 | unzip model.zip |
||
| 57 | ``` |
||
| 58 | |||
| 59 | - **input**: `shape = (N, 4096, 12)`. The input tensor should contain the `4096` points of the ECG tracings |
||
| 60 | sampled at `400Hz` (i.e., a signal of approximately 10 seconds). Both in the training and in the test set, when the |
||
| 61 | signal was not long enough, we filled the signal with zeros, so 4096 points were attained. The last dimension of the |
||
| 62 | tensor contains points of the 12 different leads. The leads are ordered in the following order: |
||
| 63 | `{DI, DII, DIII, AVR, AVL, AVF, V1, V2, V3, V4, V5, V6}`. All signal are represented as |
||
| 64 | 32 bits floating point numbers at the scale 1e-4V: so if the signal is in V it should be multiplied by |
||
| 65 | 1000 before feeding it to the neural network model. |
||
| 66 | |||
| 67 | |||
| 68 | - **output**: `shape = (N, 6)`. Each entry contains a probability between 0 and 1, and can be understood as the |
||
| 69 | probability of a given abnormality to be present. The abnormalities it predicts are **(in that order)**: 1st degree AV block(1dAVb), |
||
| 70 | right bundle branch block (RBBB), left bundle branch block (LBBB), sinus bradycardia (SB), atrial fibrillation (AF), |
||
| 71 | sinus tachycardia (ST). The abnormalities are not mutually exclusive, so the probabilities do not necessarily |
||
| 72 | sum to one. |
||
| 73 | |||
| 74 | 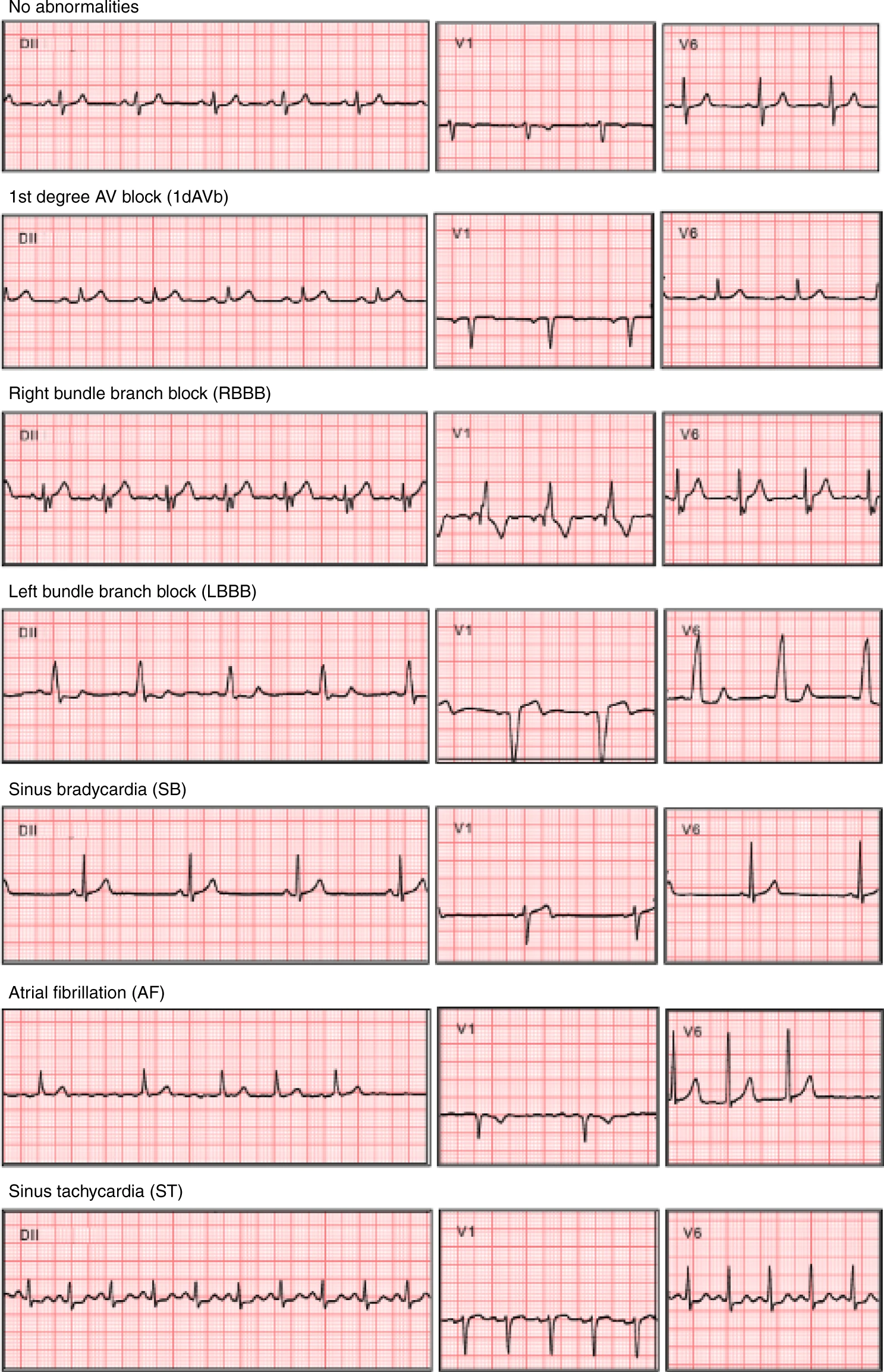 |
||
| 75 | |||
| 76 | ## Datasets |
||
| 77 | |||
| 78 | - The testing dataset described in the paper can be downloaded in: |
||
| 79 | [doi: 10.5281/zenodo.3625006](https://doi.org/10.5281/zenodo.3625006). There is also a mirror |
||
| 80 | dropbox link [here](https://www.dropbox.com/s/p3vd3plcbu9sf1o/data.zip?dl=0). |
||
| 81 | Using the command line: |
||
| 82 | ``` |
||
| 83 | wget https://www.dropbox.com/s/p3vd3plcbu9sf1o/data.zip?dl=0 -O data.zip |
||
| 84 | unzip data.zip |
||
| 85 | ``` |
||
| 86 | - Part of the training data (the CODE-15\% dataset) is openly available in: [doi: 10.5281/zenodo.4916206 ](https://doi.org/10.5281/zenodo.4916206). |
||
| 87 | - The full CODE dataset that was used for training is available upon request for research purposes: [doi: 10.17044/scilifelab.15169716](https://doi.org/10.17044/scilifelab.15169716) |
||
| 88 | |||
| 89 | ## Scripts |
||
| 90 | |||
| 91 | - ``train.py``: Script for training the neural network. To train the neural network run: |
||
| 92 | ```bash |
||
| 93 | $ python train.py PATH_TO_HDF5 PATH_TO_CSV |
||
| 94 | ``` |
||
| 95 | Pre-trained models obtained using such script can be downloaded from [here](https://doi.org/10.5281/zenodo.3625017) |
||
| 96 | |||
| 97 | |||
| 98 | - ``predict.py``: Script for generating the neural network predictions on a given dataset. |
||
| 99 | ```bash |
||
| 100 | $ python predict.py PATH_TO_HDF5_ECG_TRACINGS PATH_TO_MODEL --ouput_file PATH_TO_OUTPUT_FILE |
||
| 101 | ``` |
||
| 102 | The folder `./dnn_predicts` contain the output obtained by applying this script to the models available in |
||
| 103 | [here](https://doi.org/10.5281/zenodo.3625017) to make the predictions on tracings from |
||
| 104 | [this test dataset](https://doi.org/10.5281/zenodo.3625006). |
||
| 105 | |||
| 106 | |||
| 107 | - ``generate_figures_and_tables.py``: Generate figures and tables from the paper "Automatic Diagnosis o |
||
| 108 | the Short-Duration12-Lead ECG using a Deep Neural Network". Make sure to execute the script from the root folder, |
||
| 109 | so all relative paths are correct. So first run: |
||
| 110 | ``` |
||
| 111 | $ cd /path/to/automatic-ecg-diagnosis |
||
| 112 | ``` |
||
| 113 | Then the script |
||
| 114 | ```bash |
||
| 115 | $ python generate_figures_and_tables.py |
||
| 116 | ``` |
||
| 117 | It should generate the tables and figure in the folder `outputs/` |
||
| 118 | |||
| 119 | - ``model.py``: Auxiliary module that defines the architecture of the deep neural network. |
||
| 120 | To print a summary of the model layers run: |
||
| 121 | ```bash |
||
| 122 | $ python model.py |
||
| 123 | ``` |

 Datasets
Datasets
 Models
Models