Downloads: 1
Diff of
/README.md
[000000]
..
[c5b8c2]
Switch to unified view
| a | b/README.md | ||
|---|---|---|---|
| 1 | # ECG_Heartbeat_Classification |
||
| 2 | |||
| 3 | Description of the approach : https://blog.goodaudience.com/heartbeat-classification-detecting-abnormal-heartbeats-and-heart-diseases-from-ecgs-913449c2665 |
||
| 4 | |||
| 5 | Requirement : Keras, tensorflow, numpy |
||
| 6 | |||
| 7 | # Heartbeat Classification : Detecting abnormal heartbeats and heart diseases from |
||
| 8 | ECGs |
||
| 9 | |||
| 10 | 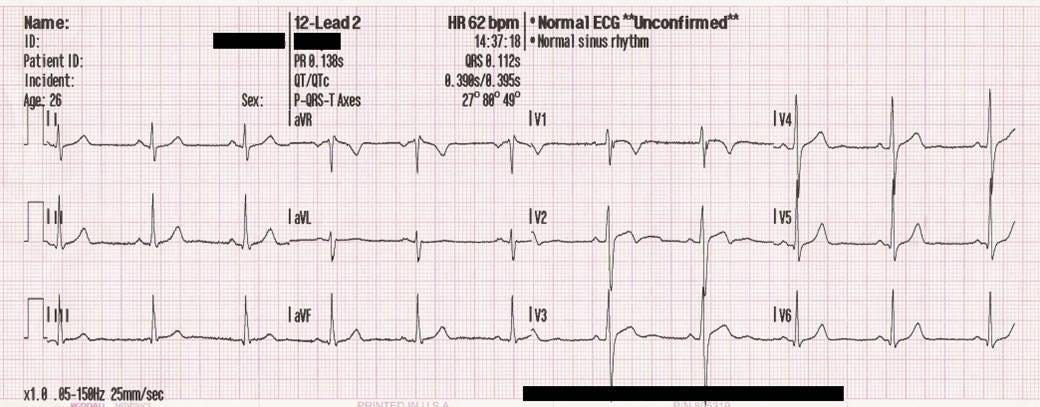 |
||
| 11 | <span class="figcaption_hack">Figure 1 : |
||
| 12 | [https://en.wikipedia.org/wiki/Electrocardiography](https://en.wikipedia.org/wiki/Electrocardiography)</span> |
||
| 13 | |||
| 14 | An ECG is a 1D signal that is the result of recording the electrical activity of |
||
| 15 | the heart using an electrode. It is one of the tool that cardiologists use to |
||
| 16 | diagnose heart anomalies and diseases. |
||
| 17 | |||
| 18 | In this blog post we are going to use an [annotated |
||
| 19 | dataset](https://www.kaggle.com/shayanfazeli/heartbeat) of heartbeats already |
||
| 20 | preprocessed by the authors of [this paper](https://arxiv.org/abs/1805.00794) to |
||
| 21 | see if we can train a model to detect abnormal heartbeats. |
||
| 22 | |||
| 23 | ### Dataset |
||
| 24 | |||
| 25 | The original datasets used are [the MIT-BIH Arrhythmia |
||
| 26 | Dataset](https://www.physionet.org/physiobank/database/mitdb/) and [The PTB |
||
| 27 | Diagnostic ECG Database](https://www.physionet.org/physiobank/database/ptbdb/) |
||
| 28 | that were preprocessed by [1] based on the methodology described in III.A of the |
||
| 29 | paper in order to end up with samples of a single heartbeat each and normalized |
||
| 30 | amplitudes as : |
||
| 31 | |||
| 32 | 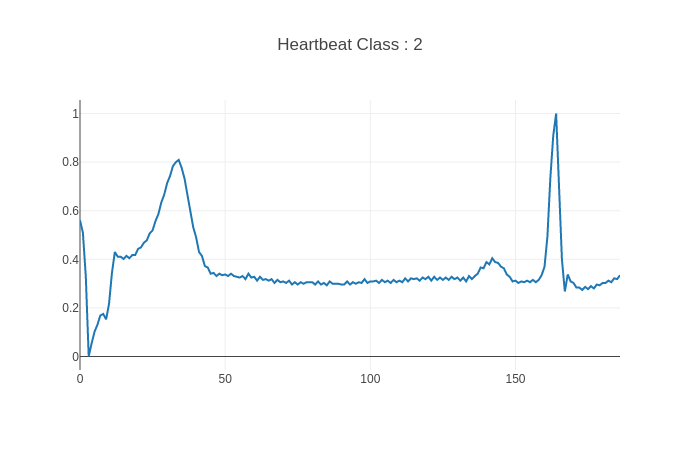 |
||
| 33 | <span class="figcaption_hack">Figure 2 : Example of preprocessed sample from the MIT-BIH dataset</span> |
||
| 34 | |||
| 35 | MIT-BIH Arrhythmia dataset : |
||
| 36 | |||
| 37 | * Number of Categories: 5 |
||
| 38 | * Number of Samples: 109446 |
||
| 39 | * Sampling Frequency: 125Hz |
||
| 40 | * Data Source: Physionet’s MIT-BIH Arrhythmia Dataset |
||
| 41 | * Classes: [’N’: 0, ‘S’: 1, ‘V’: 2, ‘F’: 3, ‘Q’: 4] |
||
| 42 | |||
| 43 | The PTB Diagnostic ECG Database |
||
| 44 | |||
| 45 | * Number of Samples: 14552 |
||
| 46 | * Number of Categories: 2 ( Normal vs Abnomal) |
||
| 47 | * Sampling Frequency: 125Hz |
||
| 48 | * Data Source: Physionet’s PTB Diagnostic Database |
||
| 49 | |||
| 50 | The published preprocessed version of the MIT-BIH dataset does not fit the |
||
| 51 | description that authors provided of it in their paper as the former is heavily |
||
| 52 | unbalanced while the latter is not. This made it so my results are not directly |
||
| 53 | comparable to theirs. I sent the authors an email to have the same split as them |
||
| 54 | and I’ll update my results if I get a reply. A similar issue exists for the PTB |
||
| 55 | dataset. |
||
| 56 | |||
| 57 | ### Model |
||
| 58 | |||
| 59 | Similar to [1] I use a neural network based on 1D convolutions but without the |
||
| 60 | residual blocks : |
||
| 61 | |||
| 62 | <span class="figcaption_hack">Figure 3 : Keras model</span> |
||
| 63 | |||
| 64 | Code : |
||
| 65 | |||
| 66 | ### Results |
||
| 67 | |||
| 68 | MIT-BIH Arrhythmia dataset : |
||
| 69 | |||
| 70 | * Accuracy : **98.5** |
||
| 71 | * F1 score : **91.5** |
||
| 72 | |||
| 73 | The PTB Diagnostic ECG Database |
||
| 74 | |||
| 75 | * Accuracy : **98.3** |
||
| 76 | * F1 score : **98.8** |
||
| 77 | |||
| 78 | ### Transferring representations |
||
| 79 | |||
| 80 | Since the PTB dataset is much smaller than the MIT-BIH dataset we can try and |
||
| 81 | see if the representations learned from MIT-BIH dataset can generalize and be |
||
| 82 | useful to the PTB dataset and improve the performance. |
||
| 83 | |||
| 84 | This can be done by loading the weights learned in MIT-BIH as initial point of |
||
| 85 | training the PTB model. |
||
| 86 | |||
| 87 | From Scratch : |
||
| 88 | |||
| 89 | * Accuracy : **98.3** |
||
| 90 | * F1 score :** 98.8** |
||
| 91 | |||
| 92 | Freezing the Convolution Layer and Training the Fully connected ones : |
||
| 93 | |||
| 94 | * Accuracy : **95.6** |
||
| 95 | * F1 score : **96.9** |
||
| 96 | |||
| 97 | Training all layers : |
||
| 98 | |||
| 99 | * Accuracy : **99.2** |
||
| 100 | * F1 score : **99.4** |
||
| 101 | |||
| 102 | We can see the freezing the first layers does not work very well. But if we |
||
| 103 | initialize the weights with those learned on MIT-BIH and train all layers we are |
||
| 104 | able to improve the performance compared to training from scratch. |
||
| 105 | |||
| 106 | Code to reproduce the results is available at : |
||
| 107 | [https://github.com/CVxTz/ECG_Heartbeat_Classification](https://github.com/CVxTz/ECG_Heartbeat_Classification) |
||
| 108 |

 Datasets
Datasets
 Models
Models