Downloads: 1
Switch to unified view
| a/README.md | b/README.md | ||
|---|---|---|---|
| 1 | # scDEC |
1 | # scDEC |
| 2 | 2 | ||
| 3 | [](https://zenodo.org/badge/latestdoi/286327774) |
3 | [](https://zenodo.org/badge/latestdoi/286327774) |
| 4 | 4 | ||
| 5 |  |
5 | 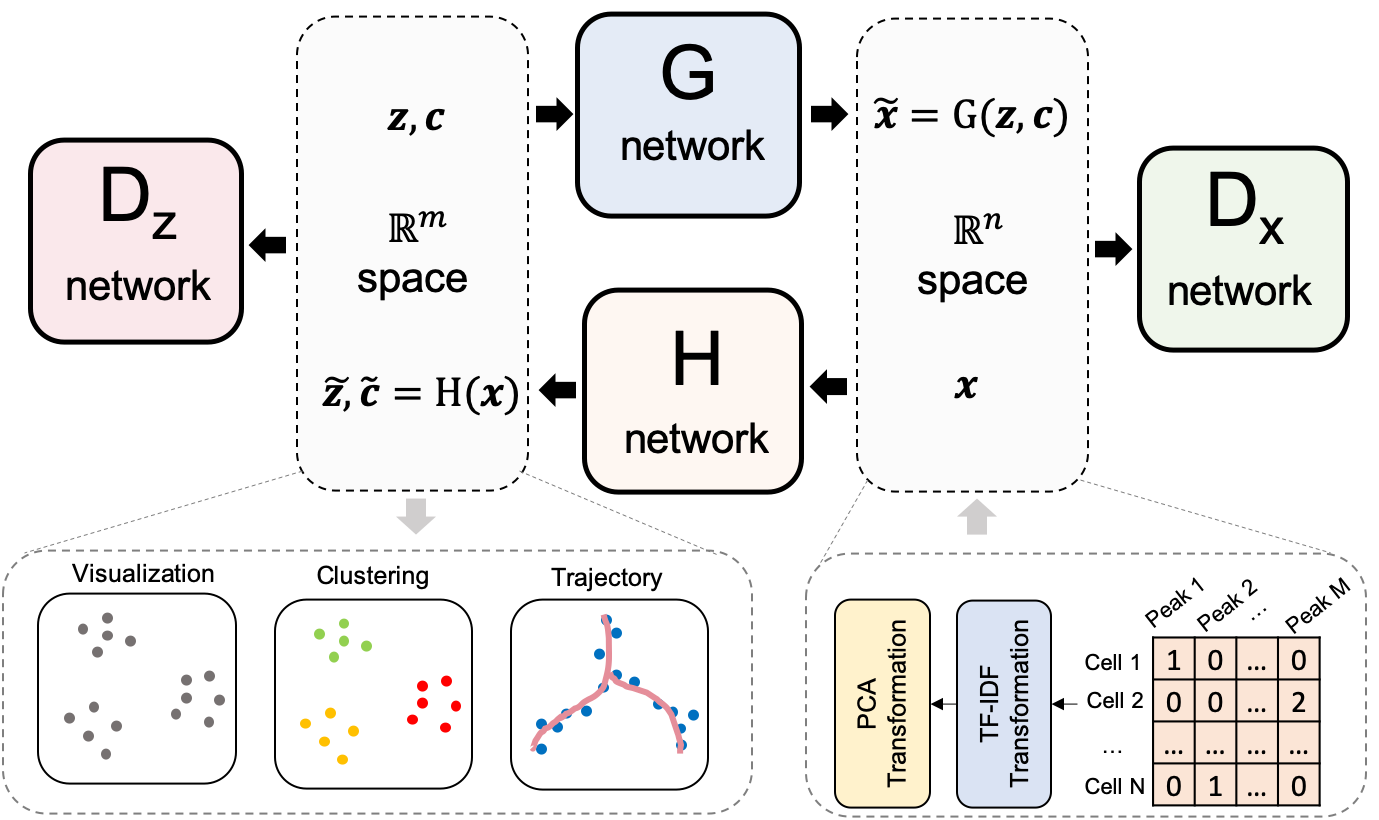 |
| 6 | 6 | ||
| 7 | scDEC is a computational tool for single cell ATAC-seq data analysis with deep generative neural networks. scDEC enables simultaneously learning the deep embedding and clustering of the cells in an unsupervised manner. scDEC is also applicable to multi-modal single cell data. We tested it on the PBMC paired data (scRNA-seq and scATAC-seq) from 10x Genomics (see Tutorials). |
7 | scDEC is a computational tool for single cell ATAC-seq data analysis with deep generative neural networks. scDEC enables simultaneously learning the deep embedding and clustering of the cells in an unsupervised manner. scDEC is also applicable to multi-modal single cell data. We tested it on the PBMC paired data (scRNA-seq and scATAC-seq) from 10x Genomics (see Tutorials). |
| 8 | 8 | ||
| 9 | ## Recent News |
9 | ## Recent News |
| 10 | 10 | ||
| 11 | An modified version of scDEC won the first place in [NeurIPS 2021 Multimodal Single-Cell Data Integration competition](https://openproblems.bio/neurips_2021/) two Joint Embedding tasks. |
11 | An modified version of scDEC won the first place in [NeurIPS 2021 Multimodal Single-Cell Data Integration competition](https://openproblems.bio/neurips_2021/) two Joint Embedding tasks. |
| 12 | 12 | ||
| 13 | ## Requirements |
13 | ## Requirements
|
| 14 | - TensorFlow==1.13.1 |
14 | - TensorFlow==1.13.1
|
| 15 | - Scikit-learn==0.19.0 |
15 | - Scikit-learn==0.19.0
|
| 16 | - Python==2.7 |
16 | - Python==2.7 |
| 17 | 17 | ||
| 18 | ## Installation |
18 | ## Installation
|
| 19 | Download scDEC by |
19 | Download scDEC by
|
| 20 | ```shell |
20 | ```shell
|
| 21 | git clone https://github.com/kimmo1019/scDEC |
21 | git clone https://github.com/kimmo1019/scDEC
|
| 22 | ``` |
22 | ```
|
| 23 | Installation has been tested in a Linux platform with Python2.7. GPU is recommended for accelerating the training process. |
23 | Installation has been tested in a Linux platform with Python2.7. GPU is recommended for accelerating the training process. |
| 24 | 24 | ||
| 25 | ## Instructions |
25 | ## Instructions |
| 26 | 26 | ||
| 27 | This section provides instructions on how to run scDEC with scATAC-seq datasets. One can also refer to [Codeocean platform](https://codeocean.com/capsule/0746056) and click `Reproducible Run` on the right. The embedding and clustering results of several datasets will be shown on the right panel. |
27 | This section provides instructions on how to run scDEC with scATAC-seq datasets. One can also refer to [Codeocean platform](https://codeocean.com/capsule/0746056) and click `Reproducible Run` on the right. The embedding and clustering results of several datasets will be shown on the right panel. |
| 28 | 28 | ||
| 29 | ### Data preparation |
29 | ### Data preparation |
| 30 | 30 | ||
| 31 | Several scATAC-seq datasets have been prepared as the input of scDEC model. These datasets can be downloaded from the [zenode repository](https://zenodo.org/record/3984189#.XzDpJRNKhTY). Uncompress the `datasets.tar.gz` in `datasets` folder then each dataset will have its own subfolder. Each dataset will contain two major files, which denote raw read count matrix (`sc_mat.txt`) and cell label (`label.txt`), respectively. The first column of `sc_mat.txt` represents the peaks information. |
31 | Several scATAC-seq datasets have been prepared as the input of scDEC model. These datasets can be downloaded from the [zenode repository](https://zenodo.org/record/3984189#.XzDpJRNKhTY). Uncompress the `datasets.tar.gz` in `datasets` folder then each dataset will have its own subfolder. Each dataset will contain two major files, which denote raw read count matrix (`sc_mat.txt`) and cell label (`label.txt`), respectively. The first column of `sc_mat.txt` represents the peaks information. |
| 32 | 32 | ||
| 33 | ### Model training |
33 | ### Model training |
| 34 | 34 | ||
| 35 | scDEC is an unsupervised learning model for analyzing scATAC-seq data. One can run |
35 | scDEC is an unsupervised learning model for analyzing scATAC-seq data. One can run |
| 36 | 36 | ||
| 37 | ```python |
37 | ```python
|
| 38 | python main_clustering.py --data [dataset] --K [nb_of_clusters] --dx [x_dim] --dy [y_dim] --train [is_train] |
38 | python main_clustering.py --data [dataset] --K [nb_of_clusters] --dx [x_dim] --dy [y_dim] --train [is_train]
|
| 39 | [dataset] - the name of the dataset (e.g.,Splenocyte) |
39 | [dataset] - the name of the dataset (e.g.,Splenocyte)
|
| 40 | [nb_of_clusters] - the number of clusters (e.g., 6) |
40 | [nb_of_clusters] - the number of clusters (e.g., 6)
|
| 41 | [x_dim] - the dimension of Gaussian distribution |
41 | [x_dim] - the dimension of Gaussian distribution
|
| 42 | [y_dim] - the dimension of PCA (defalt: 20) |
42 | [y_dim] - the dimension of PCA (defalt: 20)
|
| 43 | [is_train] - indicate training from scratch or using pretrained model |
43 | [is_train] - indicate training from scratch or using pretrained model |
| 44 | 44 | ||
| 45 | ``` |
45 | ```
|
| 46 | For an example, one can run `CUDA_VISIBLE_DEVICES=0 python main_clustering.py --data Splenocyte --K 12 --dx 8 --dy 20` to cluster the scATAC-seq data with pretrained model. Note that the dimension of the embedding should be `K+x_dim` |
46 | For an example, one can run `CUDA_VISIBLE_DEVICES=0 python main_clustering.py --data Splenocyte --K 12 --dx 8 --dy 20` to cluster the scATAC-seq data with pretrained model. Note that the dimension of the embedding should be `K+x_dim` |
| 47 | 47 | ||
| 48 | Or one can run `CUDA_VISIBLE_DEVICES=0 python main_clustering.py --data Splenocyte --K 12 --dx 8 --dy 20 --train True` to train the model from scratch. |
48 | Or one can run `CUDA_VISIBLE_DEVICES=0 python main_clustering.py --data Splenocyte --K 12 --dx 8 --dy 20 --train True` to train the model from scratch. |
| 49 | 49 | ||
| 50 | ### Model evaluation |
50 | ### Model evaluation |
| 51 | 51 | ||
| 52 | If the pretrained model was used, the clustering results in the last step will be directly saved in `results/[dataset]/data_pre.npz` where `dataset` is the name of the scATAC-seq dataset. Note that `data_pre.npz` or `data_at_xxx.npz` contains the predictions from the H network. The first part denotes the embeddings and the second part denotes the inferred one-hot label where one can use `np.argmax` function to get the cluster label. |
52 | If the pretrained model was used, the clustering results in the last step will be directly saved in `results/[dataset]/data_pre.npz` where `dataset` is the name of the scATAC-seq dataset. Note that `data_pre.npz` or `data_at_xxx.npz` contains the predictions from the H network. The first part denotes the embeddings and the second part denotes the inferred one-hot label where one can use `np.argmax` function to get the cluster label. |
| 53 | 53 | ||
| 54 | Then one can run `python eval.py --data [dataset]` to analyze the clustering results. |
54 | Then one can run `python eval.py --data [dataset]` to analyze the clustering results.
|
| 55 | For an example, one can run `python eval.py --data Splenocyte` |
55 | For an example, one can run `python eval.py --data Splenocyte` |
| 56 | 56 | ||
| 57 | The t-SNE visualization plot of latent features (`scDEC_embedding.png`), latent feature matrix (`scDEC_embedding.csv`), inferred cluster label (`scDEC_cluster.txt`) will be saved in the `results/[dataset]` folder. |
57 | The t-SNE visualization plot of latent features (`scDEC_embedding.png`), latent feature matrix (`scDEC_embedding.csv`), inferred cluster label (`scDEC_cluster.txt`) will be saved in the `results/[dataset]` folder. |
| 58 | 58 | ||
| 59 | 59 | ||
| 60 | If scDEC model was trained from scratch, the results will be marked by a unique timestamp YYYYMMDD_HHMMSS. This timestamp records the exact time when you run the script. The outputs from the training includes: |
60 | If scDEC model was trained from scratch, the results will be marked by a unique timestamp YYYYMMDD_HHMMSS. This timestamp records the exact time when you run the script. The outputs from the training includes: |
| 61 | 61 | ||
| 62 | 1) `log` files and predicted assignmemnts `data_at_xxx.npz` (xxx denotes different epoch) can be found at folder `results/[dataset]/YYYYMMDD_HHMMSS_x_dim=8_y_dim=20_alpha=10.0_beta=10.0_ratio=0.2`. |
62 | 1) `log` files and predicted assignmemnts `data_at_xxx.npz` (xxx denotes different epoch) can be found at folder `results/[dataset]/YYYYMMDD_HHMMSS_x_dim=8_y_dim=20_alpha=10.0_beta=10.0_ratio=0.2`.
|
| 63 | 63 |
|
|
| 64 | 2) Model weights will be saved at folder `checkpoint/YYYYMMDD_HHMMSS_x_dim=8_y_dim=20_alpha=10.0_beta=10.0_ratio=0.2`. |
64 | 2) Model weights will be saved at folder `checkpoint/YYYYMMDD_HHMMSS_x_dim=8_y_dim=20_alpha=10.0_beta=10.0_ratio=0.2`.
|
| 65 | 65 |
|
|
| 66 | 3) The training loss curves were recorded at folder `graph/YYYYMMDD_HHMMSS_x_dim=8_y_dim=20_alpha=10.0_beta=10.0_ratio=0.2`, which can be visualized using TensorBoard. |
66 | 3) The training loss curves were recorded at folder `graph/YYYYMMDD_HHMMSS_x_dim=8_y_dim=20_alpha=10.0_beta=10.0_ratio=0.2`, which can be visualized using TensorBoard. |
| 67 | 67 | ||
| 68 | Next, one can run |
68 | Next, one can run
|
| 69 | 69 |
|
|
| 70 | ```python |
70 | ```python
|
| 71 | python eval.py --data [dataset] --timestamp [timestamp] --epoch [epoch] --train [is_train] |
71 | python eval.py --data [dataset] --timestamp [timestamp] --epoch [epoch] --train [is_train]
|
| 72 | [dataset] - the name of the dataset (e.g.,Splenocyte) |
72 | [dataset] - the name of the dataset (e.g.,Splenocyte)
|
| 73 | [timestamp] - the timestamp of the experiment you ran |
73 | [timestamp] - the timestamp of the experiment you ran
|
| 74 | [epoch] - specify to use the results of which epoch (it can be ignored) |
74 | [epoch] - specify to use the results of which epoch (it can be ignored)
|
| 75 | [is_train] - indicate training from scratch |
75 | [is_train] - indicate training from scratch
|
| 76 | ``` |
76 | ``` |
| 77 | 77 | ||
| 78 | E.g., `python eval.py --data All_blood --timestamp 20200910_143208 --train True` |
78 | E.g., `python eval.py --data All_blood --timestamp 20200910_143208 --train True` |
| 79 | 79 | ||
| 80 | The t-SNE visualization plot of latent features (`scDEC_embedding.png`), latent feature matrix (`scDEC_embedding.csv`), inferred cluster label (`scDEC_cluster.txt`) will be saved in the same `results` folder as 1). |
80 | The t-SNE visualization plot of latent features (`scDEC_embedding.png`), latent feature matrix (`scDEC_embedding.csv`), inferred cluster label (`scDEC_cluster.txt`) will be saved in the same `results` folder as 1). |
| 81 | 81 | ||
| 82 | 82 | ||
| 83 | ### Analyzing scATAC dataset without label |
83 | ### Analyzing scATAC dataset without label |
| 84 | 84 | ||
| 85 | One can also use scDEC to analyze custome scATAC-seq dataset, especially the label is unknown. First, the users should prepare raw read count matrix (`sc_mat.txt`) under the folder `datasets/[NAME]`. `[NAME]` denotes the dataset name. |
85 | One can also use scDEC to analyze custome scATAC-seq dataset, especially the label is unknown. First, the users should prepare raw read count matrix (`sc_mat.txt`) under the folder `datasets/[NAME]`. `[NAME]` denotes the dataset name. |
| 86 | 86 | ||
| 87 | Second, one can run the following command: |
87 | Second, one can run the following command: |
| 88 | 88 | ||
| 89 | ```python |
89 | ```python
|
| 90 | python main_clustering.py --data [dataset] --K [nb_of_clusters] --dx [x_dim] --dy [y_dim] --train [is_train] --no_label |
90 | python main_clustering.py --data [dataset] --K [nb_of_clusters] --dx [x_dim] --dy [y_dim] --train [is_train] --no_label
|
| 91 | [dataset] - the name of the dataset (e.g.,Mydataset) |
91 | [dataset] - the name of the dataset (e.g.,Mydataset)
|
| 92 | [nb_of_clusters] - the number of clusters (e.g., 6) |
92 | [nb_of_clusters] - the number of clusters (e.g., 6)
|
| 93 | [x_dim] - the dimension of latent space (continous part) |
93 | [x_dim] - the dimension of latent space (continous part)
|
| 94 | [y_dim] - the dimension of PCA (defalt: 20) |
94 | [y_dim] - the dimension of PCA (defalt: 20)
|
| 95 | [is_train] - indicate training from scratch |
95 | [is_train] - indicate training from scratch
|
| 96 | ``` |
96 | ``` |
| 97 | 97 | ||
| 98 | For an example, one can run `CUDA_VISIBLE_DEVICES=0 python main_clustering.py --data Mydataset --K 10 --dx 5 --dy 20 --train True --no_label` to clustering custom dataset. |
98 | For an example, one can run `CUDA_VISIBLE_DEVICES=0 python main_clustering.py --data Mydataset --K 10 --dx 5 --dy 20 --train True --no_label` to clustering custom dataset. |
| 99 | 99 | ||
| 100 | Then one can run `python eval.py --data Mydataset --timestamp YYYYMMDD_HHMMSS --epoch epoch --no_label`. Nota time the timestamp `YYYYMMDD_HHMMSS` (for training) and epoch/batch index `epoch` (the last training epoch/batch index is recommended) should be provided. The clustering results (cluster assignments) will be saved in the `results/Mydataset/YYYYMMDD_HHMMSS_xxx` folder. |
100 | Then one can run `python eval.py --data Mydataset --timestamp YYYYMMDD_HHMMSS --epoch epoch --no_label`. Nota time the timestamp `YYYYMMDD_HHMMSS` (for training) and epoch/batch index `epoch` (the last training epoch/batch index is recommended) should be provided. The clustering results (cluster assignments) will be saved in the `results/Mydataset/YYYYMMDD_HHMMSS_xxx` folder. |
| 101 | 101 | ||
| 102 | 102 | ||
| 103 | 103 | ||
| 104 | ## Tutorial |
104 | ## Tutorial |
| 105 | 105 | ||
| 106 | [Tutorial Splenocyte](https://github.com/kimmo1019/scDEC/wiki/Splenocyte) Run scDEC on Splenocyte dataset (3166 cells) |
106 | [Tutorial Splenocyte](https://github.com/kimmo1019/scDEC/wiki/Splenocyte) Run scDEC on Splenocyte dataset (3166 cells) |
| 107 | 107 | ||
| 108 | [Tutorial Full mouse atlas](https://github.com/kimmo1019/scDEC/wiki/Full-Mouse-atlas) Run scDEC on full Mouse atlas dataset (81173 cells) |
108 | [Tutorial Full mouse atlas](https://github.com/kimmo1019/scDEC/wiki/Full-Mouse-atlas) Run scDEC on full Mouse atlas dataset (81173 cells) |
| 109 | 109 | ||
| 110 | [Tutorial PBMC10k paired data ](https://github.com/kimmo1019/scDEC/wiki/PBMC10k) Run scDEC on PBMC data, which contains around 10k cells with both scRNA-seq and scATAC-seq (labels were manually annotated from 10x Genomic R&D group) |
110 | [Tutorial PBMC10k paired data ](https://github.com/kimmo1019/scDEC/wiki/PBMC10k) Run scDEC on PBMC data, which contains around 10k cells with both scRNA-seq and scATAC-seq (labels were manually annotated from 10x Genomic R&D group)
|
| 111 | 111 |
|
|
| 112 | ## Contact |
112 | ## Contact |
| 113 | 113 | ||
| 114 | Also Feel free to open an issue in Github or contact `liuqiao@stanford.edu` if you have any problem in running scDEC. |
114 | Also Feel free to open an issue in Github or contact `liuqiao@stanford.edu` if you have any problem in running scDEC. |
| 115 | 115 | ||
| 116 | ## License |
116 | ## License |
| 117 | 117 | ||
| 118 | This project is licensed under the MIT License - see the LICENSE.md file for details |
118 | This project is licensed under the MIT License - see the LICENSE.md file for details
|

 Datasets
Datasets
 Models
Models