90 lines (58 with data), 24.0 kB
Thematic analysis of UK funding for AI and genomics research
Previously, we used the categories that Gateway to Research documents have been tagged with and the text in their abstracts to identify projects related to AI and genomics. We also extracted information about documents in the broader AI and genomics categories, which we will use as baselines.
Here, we develop a bottom-up typology of projects based on a semantic clustering of their abstracts and analyse their evolution over time. This is a prototype analysis that could be deployed for thematic analyses in other data sources we are using in the project.
Methodology
Our starting point are ca. 1,200 projects including 257 projects that have identified as belonging to AI and genomics, and a randomly selected group of "AI" and "genomics" projects providing a baseline for analysis.
We have created vector representations of these projects' abstracts using SPECTER, an open language model trained on a scientific corpus. This results on a 768-dimensional representation of each abstract (project) capturing its position in semantic space.
We reduce the dimensionality of these data using UMAP (Uniform Manifold Approximation and Projection), an algorithm that projects high-dimensional data into fewer (2 in this case) dimensions while retaining the local structure of the larger and more complex dataset. @gtr_umap presents the results, using color to distinguish between AI and genomics projects and projects in the broader "AI" and "genomics" categories.
 {#fig:gtr_umap}
{#fig:gtr_umap}
We note that AI and genomics projects (orange) tend to appear close to clusters of genomics papers (red), suggesting that they mostly comprise applications of AI techniques to existing areas of genomics. We also find a small number of AI projects clustering with genomics projects. When we inspect their titles, we find that they are in areas adjacent to genomics such as drug development and biophysics. This suggests that our semantic clustering is capturing the content of abstracts, and that it could be used to identify false negatives in the data i.e. bona fide AI and genomics projects that were not captured by our category and keyword-based search strategy. @fig:gtr_cluster_distr presents the number of documents in each cluster and their distribution over different categories. It shows a number of clusters with dominance by AI (e.g. 42, 27, 18, 12), clusteers with a predominance of genomics projects and mixed ones, potentially suggesting the scope for deploying AI in different domains of genomics.
 {#fig:gtr_cluster_distr}
{#fig:gtr_cluster_distr}
Results
We cluster papers on a smaller number of categories using the K-Means algorith, which searches for homogeneous (closely packed) collections of elements in the 2-dimensional output of the UMAP projection. We run five instances of K-Means for different values of K (number of clusters) ranging between 5 and 100.
The boxplot in @fig:gtr_sil shows the distribution of silhouette scores (a measure of clustering goodness-of-fit i.e. the extent to which entities in a cluster are similar to each other and different fromn those outside ranging between -1 and 1, which higher values indicating better clustering) as K grows. We see an initial plateau in performance increases at around 50 and chose that value as a pragmatic compromise between cluster homogeneity and tractability (avoiding an excessive number of categories) when reporting.
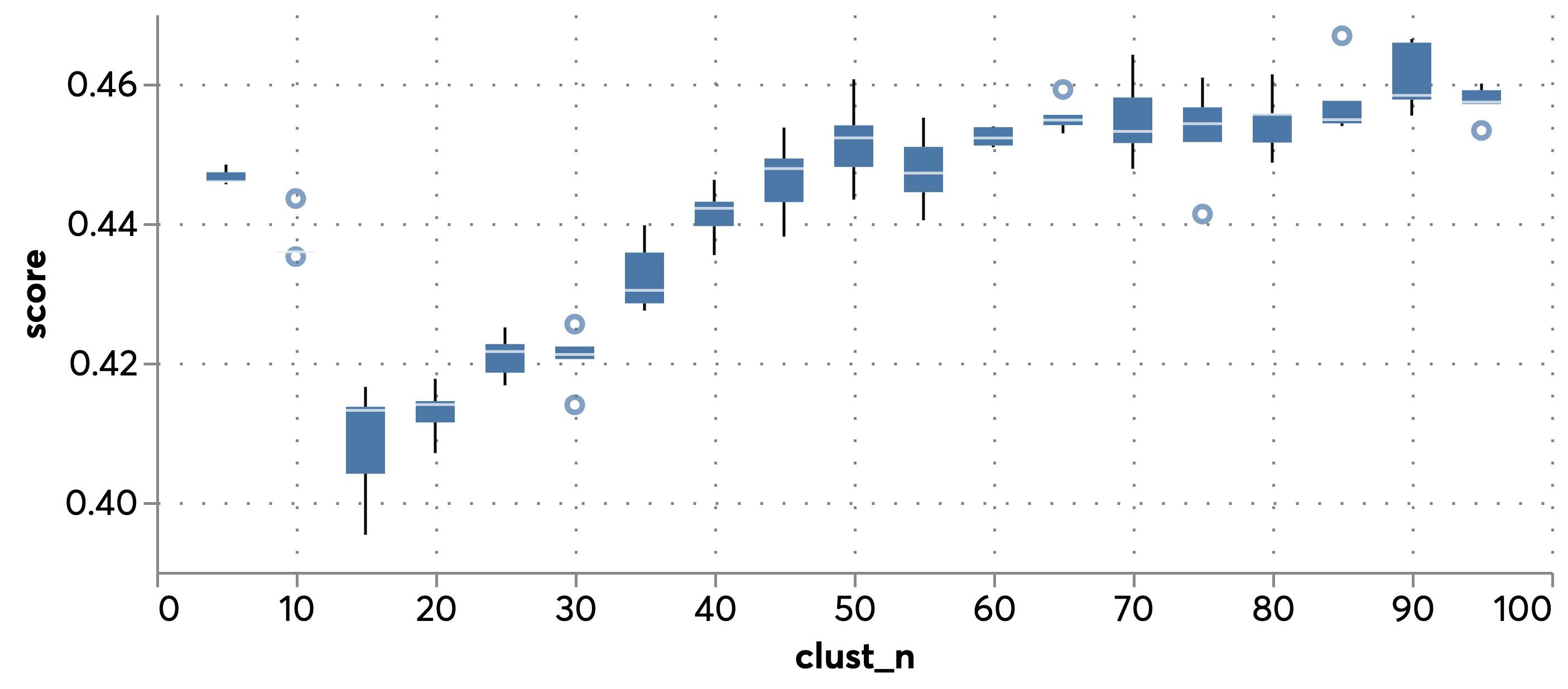 {#fig:gtr_sil}
{#fig:gtr_sil}
Cluster content
@tab:gtr_clusters presents example clusters focusing on 19 clusters with at least five AI and genomics projects. For each of these clusters, we present the titles from five randomly drawn AI and genomics projects.
| cluster id | n docs | n AI & genom | % AI & genom | examples |
|---|---|---|---|---|
| 0 | 26 | 20 | 0.769 | Precision Medicine in Crohn's Disease and Ulcerative Colitis: Predicting Disease Flare and Disease progression, Artificial intelligence to create equitable multi-ethnic polygenic risk scores that improve clinical care, Predictive analytics of integrated genomic and clinical data using machine learning and complex statistical approaches, Integrating transcriptomic and metabolomic data from people with rheumatoid arthritis to predict clinical response to drug treatment, Predictive analytics of integrated genomic and clinical data using machine learning and complex statistical approaches |
| 16 | 10 | 7 | 0.7 | A Decision Support tool for Potato Blackleg Disease (DeS-BL), A Decision Support tool for Potato Blackleg Disease (DeS-BL), New tools for predicting spread of Fusarium wilt in banana, A Decision Support tool for Potato Blackleg Disease (DeS-BL) - Blackleg, A Decision Support tool for Potato Blackleg Disease (DeS-BL) |
| 36 | 33 | 20 | 0.606 | Novel statistical techniques for joint estimation of selection and migration from time series genetic data, Why doesn't the evolution of antibiotic resistance have a larger fitness cost?, Improving Bayesian methods for estimating divergence times integrating genomic and trait data, Statistical Methods for Genomic Analysis of Species Divergences, Bayesian Inference under the Structured Coalescent Model |
| 29 | 15 | 9 | 0.6 | Exploring disentangled generative factors of cancer transcriptomes, Novel Multi-Omics Integration and Machine Learning Tools to Understand Heterogeneous Systems of Ovarian Cancers, Statistical methods for ovarian cancer diagnosis and prognosis, From single cells to populations: generalized pseudotime analysis to identify patient trajectories from cross-sectional data in cancer genomics, From single cells to populations: generalized pseudotime analysis to identify patient trajectories from cross-sectional data in cancer genomics |
| 17 | 19 | 10 | 0.526 | GREET: Generative Recombinant Enzyme Engineering for Therapeutics, Synthetic Biology Lab Automation and the Application of Artificial Intelligence, Bayesian machine learning and Self-Assembly, Design/Build/Test/Learn: novel antibiotic discovery using a plug-and-play robotic platform, Biodesign and Engineering of Functionalized Spider Silk Variants |
| 46 | 18 | 9 | 0.5 | A Multi-Scale Approach to Classifying DNA Damage Repair Deficiencies and their Therapeutic Relevance, Neurobiology of multisensory decision making in humans, Comparative Analyses and Genomic Sequence-Only Prediction of DNA Breakpoints via Gradient Boosting Machines and Deep Learning, Imputation-aided analysis of DNA methylomes, AI based approaches multi-dimensional functional genomics |
| 49 | 21 | 10 | 0.476 | Machine learning of RNA code artifacts in inflammatory diseases towards precision medicine, Deep-Learning Algorithms for Evolutionary Inferences from Genomic and Ecological Data, Deep Learning for Inference from Biological Data, Predicting dominant mutations in genetic disorders associated with the misassembly of cytoskeletal proteins, Bayesian models of expression in the transcriptome for clinical RNA-seq |
| 10 | 26 | 12 | 0.462 | i-sense: EPSRC IRC in Agile Early Warning Sensing Systems for Infectious Diseases and Antimicrobial Resistance, Genomic Epidemiology and Transmission of Campylobacter in Africa (GETcampy-Africa), Medical Mycology and Fungal Immunology, Medical Mycology and Fungal Immunology, Combatting Antimicrobial Resistance with Next-Generation Diagnostics for Sepsis |
| 38 | 25 | 11 | 0.44 | Development of a molecular barcoding tool for Plasmodium malaria, Genomic epidemiology of infectious disease outbreaks, Mapping the patterns and drivers of antibiotic use and environmental resistance in the Argentine beef industry., Using phylogenetic analysis of large-scale Next-Gen HIV sequence datasets and computer simulation to assess the impact of high-risk populations on the, Machine-learning to predict and understand the zoonotic threat of E. coli O157 isolates |
| 31 | 20 | 8 | 0.4 | Integrating circadian, neuroimaging and genetic data to investigate major depression and bipolar disorder, Leveraging the impact of diversity in neurodevelopmental disability by integrating machine learning in personalized interventions., Psychosis Immune Mechanism Stratified Medicine Study (PIMS), Integrative Omics approaches for understanding molecular pathogenesis of Dementia with Lewy bodies, Enhancing Neuroimaging Genetics through Meta-analysis (ENIGMA): Antisocial Behaviour working group |
| 47 | 23 | 9 | 0.391 | Prediction of protein-protein interaction hot spots using a combination of physics and machine learning, Machine Learning Approaches to Predict Enzyme Function, Protein Function Prediction using Machine Learning by an Enhanced Novel Support Vector Logic-based Approach, Accelerating and enhancing the PSIPRED Workbench with deep learning, Accelerating and enhancing the PSIPRED Workbench with deep learning |
| 30 | 19 | 7 | 0.368 | How will cold adapted life respond to climate change? - Using artificial intelligence to decipher life in the cold., EPSRC and MRC Centre for Doctoral Training in Systems Approaches to Biomedical Science, EMERALD - Enriching MEtagenomics Results using Artificial intelligence and Literature Data, Designing next-generation therapeutics for atopic eczema by controlling skin microbiome, Development of Computational Methods and Tools for the Mining of Novel Biocatalysts in Metagenomes |
| 24 | 23 | 8 | 0.348 | The contribution of metabolic switching to an emerging human pathogen from the genus Photorhabdus, Phase variable epigenetic control of strain fitness of starter cultures and probiotics, RNA Polymerase III in healthy ageing: consolidating the mechanisms of longevity from worms and flies to mice, RNA Polymerase III in healthy ageing: consolidating the mechanisms of longevity from worms and flies to mice, Targeting redox-active cysteine residues in protein kinases as a new therapeutic approach to ageing and age-associated diseases |
| 9 | 32 | 11 | 0.344 | Scalable causal gene network inference via genetic node ordering, Embracing multi-ethnicity in studying the genetics of smoking behaviour, Investigating the mechanisms underlying disease using multiOmics data, Identification of genomic biomarkers associated with hypertrophic cardiomyopathy (HCM) in cats, Elucidating the genetic background of rare neurological diseases: with a focus on paediatric mitochondrial disorders |
| 27 | 29 | 7 | 0.241 | Deep Learning for Behavioural Genomics, Deep Learning for Behavioural Genomics, Creating, screening, and modelling multiplexed reporters of cell state, Developing an integrative approach to phenomics for industrial, biomedical and environmental applications, Bayesian modelling for developmental systems biology |
| 4 | 31 | 7 | 0.226 | Computational Developments, Data mining and characterisation of dark metagenomic sequence data, MRes in Computational Biology, Evolutionary and thermodynamical features of musculoskeletal disease mutations is human intrinsically disordered protein regions, Bayesian Computation in Systems and Synthetic Biology |
| 20 | 27 | 6 | 0.222 | Production of genetically modified chickens resistant to major avian respiratory viral pathogens, Investigating Host and Viral Factors for Improved Design of Future Live Attenuated Vaccines for IBV, Production of genetically modified chickens resistant to important avian respiratory diseases., Combating highly pathogenic avian influenza: novel vaccination strategies, Shining a light on dense granules- biochemical, genetic and cell biological investigation of an essential but understudied compartment in malarial - |
| 14 | 31 | 6 | 0.194 | Mapping the functional gene-scape of the oceans under conditions of global change, The role of sponge competition in cold-water coral reefs - ROV video analyses and secondary metabolite profiling, Big-data analysis tools for bridging the gap between omics and earth system science, Application of Computational Methods and Multiomic Techniques to the Analysis of Marine Life, Mapping Microbial Community Structure and Function in Enhanced Biological Phosphorus Systems |
| 1 | 38 | 7 | 0.184 | Landscape genomics of Bornean water buffalos, Fighting Infection and AMR in broiler farming: AI, omics and smart sensing for diagnostics, treatment selection and gut microbiome improvement, Improving livestock production through high-throughput identification of functional regulatory variation, Design of breeding programs to improve honeybee health and production, Microbiome metagenomics, senescence and mortality in an island population of birds |
- Example projects from the largest and more significant AI and genomics clusters {#tab:gtr_clusters}
Our clustering seems to capture different themes / types of AI genomics in the GtR data. For example, cluster 0 is focused on the use of genomic data for precision medicine; cluster 36 uses genomic data for population analysis; cluster 29 focuses on cancer applications; cluster 17 on biotechnology applications; cluster 46 on DNA analysis; cluster 49 on RNA and mutations; cluster 38 on diseases primarily prevalent in developing countries etc.
Trend analysis
We conclude our exploration of the data with an analysis of trends in different AI and genomics topics in the Gateway to Research.
In @fig:gtr_cluster_trends, every row is a cluster of documents (focusing on those with at least 5 AI and genomics projects), the size of the squares represents the number of projects, and the colour the proportion of AI and genomics projects in that category and year. The clusters are sorted by the number of AI and genomics projects that they contain, with more AI and genomics intensive clusters towards the top.
This chart helps us to distinguish between well established research themes such as 36 (genetic analysis) and 47 (proteomics) and research themes where most of the activity has been happening recently such as 0 (predictive analyses for precision medicine), 17 (use of AI and genomics data in biotechnology and the automation of drug discovery) and 20 (which seems related to avian influenza). We also note a number of clusters with no activity in the last couple of years, such as 31 (focused on the analysis of neurological disorders and mental health issues) or 29 (cancer applications).
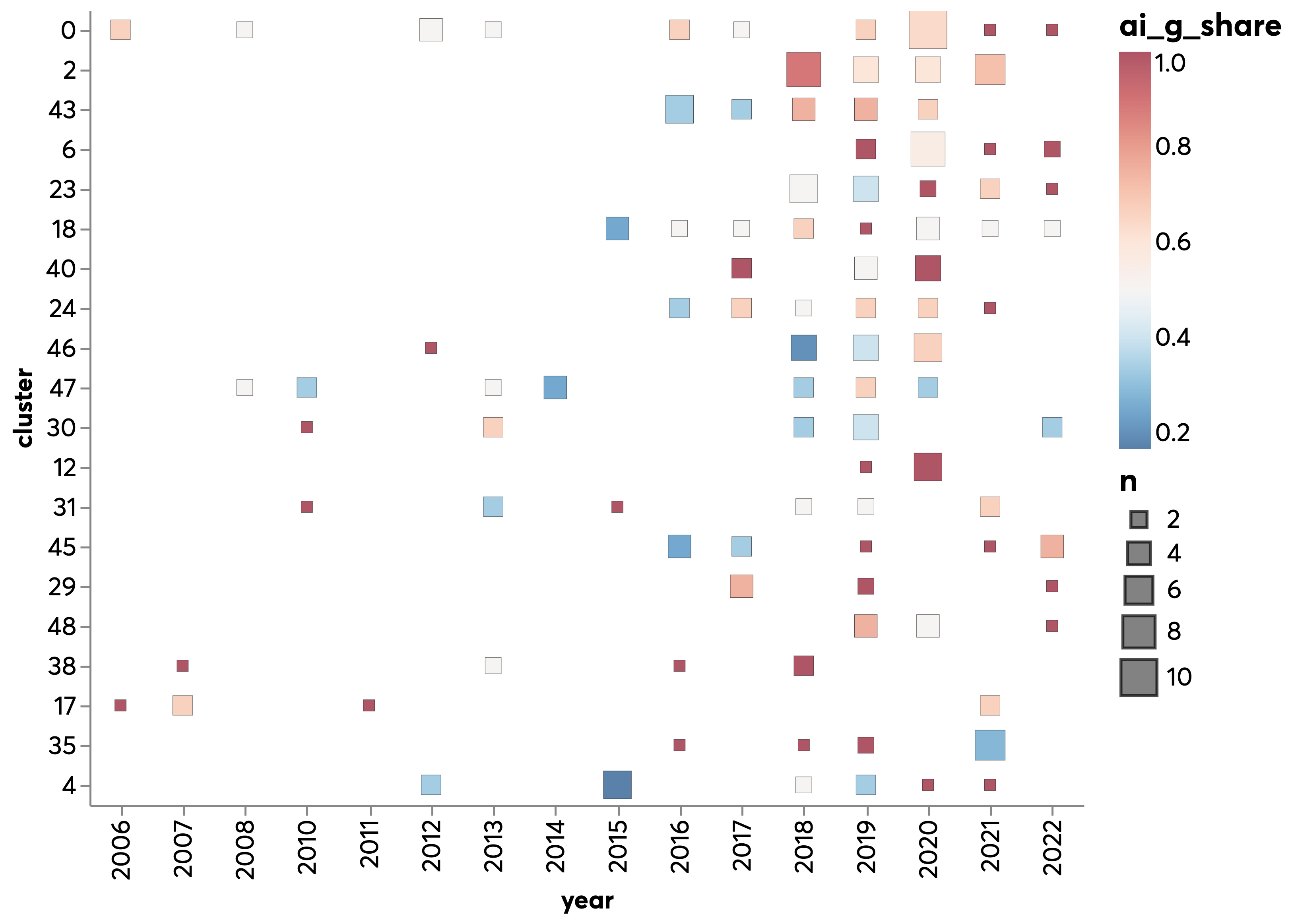 {#fig:gtr_cluster_trends)
{#fig:gtr_cluster_trends)
In @fig:gtr_cluster_trends we adapt the methodology used by Nesta in previous analyses of technological emergence to our data. For each of the clusters with more than 5 AI and genomics projects, we calculate measures of significance (% of all projects since 2020 accounted for by the cluster) and recency (% of all projects in a cluster initiated in the last three years). This seeks to help us distinguish between emerging (high recency and low significance), hot (high recency and high significance), stabilising (high significance and low recency) clusters. We present the results in @fig:gtr_emergence_scatter and @tbl:gtr_cluster_class.
 {#fig:gtr_emergence_scatter}
{#fig:gtr_emergence_scatter}
| Recency / Significance | Low | High |
|---|---|---|
| Low | Inert: 47 (proteomics / high performance computing), 24 (unclear interpretation) | Established: 0 (Predictive analysis and precision medicine), 36 (Evolutionary analyses) |
| High | Emergent: 1 (Agricultural applications), 31 (neurological disorders, mental health) | Hot: 10 (Antimicrobial resistance, immunology), 17 (Biotechnology and automation applications), 9 (causal determinants of disease) |
- Preliminary classification of clusters {#tbl:gtr_cluster_class}
We note that we were not able to tag some of the clusters in the table, suggesting that they might benefit from additional decomposition into a larger number of clusters.
Concluding remarks
We have piloted a potential strategy to segment GtR projects into clusters to analyse its emergence. Even with the current (rough, heuristic-based) clustering strategy, the results show potential for understanding recent trends in applications of AI to genomics. We conclude by highlighting limitations and extensions.
- As @fig:distr_sil shows, there is scope to improve clustering performance by extracting more clusters from the data but this would be at the expense of readibility and potentially interpretability. One option would be to focus additional clusters on heterogeneous and hard to interpret clusters.
- We are currently tagging clusters manually. It would be useful, specially if we increase the resolution of our clustering, to develop automated strategies for cluster-naming based on salient terms and phrases in clusters, text summarisation or some other strategy.
- It would be straightforward to apply vector representation -> clustering -> analysis pipeline that we have prototyped here to other data sources in the project - in particular OpenAlex publications, which use a similar vocabulary to the scientific corpus where SPECTER was trained. We note that SPECTER vectors are already available from the Semantic Scholar platform so this would not require computationally expensive vector generation on our part.
- The methodology prototyped here could be expanded to generate measures of novelty / interdisciplinarity at the document level based on the extent to which a document eccentric and hard to classify into a single cluster.

 Datasets
Datasets
 Models
Models