88 lines (61 with data), 5.5 kB
AGCN
Abstract
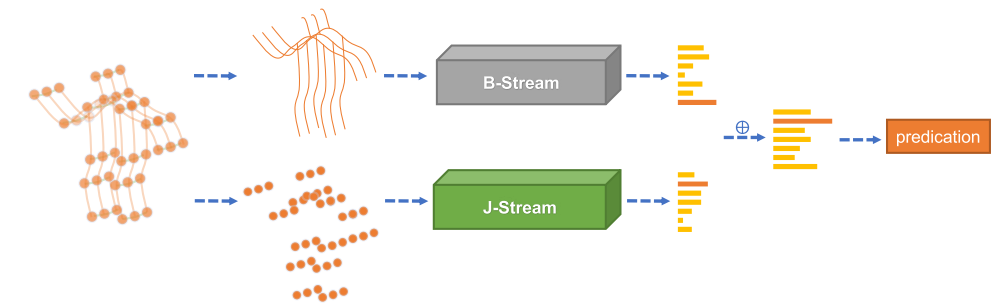
In skeleton-based action recognition, graph convolutional networks (GCNs), which model the human body skeletons as spatiotemporal graphs, have achieved remarkable performance. However, in existing GCN-based methods, the topology of the graph is set manually, and it is fixed over all layers and input samples. This may not be optimal for the hierarchical GCN and diverse samples in action recognition tasks. In addition, the second-order information (the lengths and directions of bones) of the skeleton data, which is naturally more informative and discriminative for action recognition, is rarely investigated in existing methods. In this work, we propose a novel two-stream adaptive graph convolutional network (2s-AGCN) for skeleton-based action recognition. The topology of the graph in our model can be either uniformly or individually learned by the BP algorithm in an end-to-end manner. This data-driven method increases the flexibility of the model for graph construction and brings more generality to adapt to various data samples. Moreover, a two-stream framework is proposed to model both the first-order and the second-order information simultaneously, which shows notable improvement for the recognition accuracy. Extensive experiments on the two large-scale datasets, NTU-RGBD and Kinetics-Skeleton, demonstrate that the performance of our model exceeds the state-of-the-art with a significant margin.
Citation
@inproceedings{shi2019two,
title={Two-stream adaptive graph convolutional networks for skeleton-based action recognition},
author={Shi, Lei and Zhang, Yifan and Cheng, Jian and Lu, Hanqing},
booktitle={Proceedings of the IEEE/CVF conference on computer vision and pattern recognition},
pages={12026--12035},
year={2019}
}
Model Zoo
NTU60_XSub
| config | type | gpus | backbone | Top-1 | ckpt | log | json |
|---|---|---|---|---|---|---|---|
| 2sagcn_80e_ntu60_xsub_keypoint_3d | joint | 1 | AGCN | 86.06 | ckpt | log | json |
| 2sagcn_80e_ntu60_xsub_bone_3d | bone | 2 | AGCN | 86.89 | ckpt | log | json |
Train
You can use the following command to train a model.
python tools/train.py ${CONFIG_FILE} [optional arguments]
Example: train AGCN model on joint data of NTU60 dataset in a deterministic option with periodic validation.
python tools/train.py configs/skeleton/2s-agcn/2sagcn_80e_ntu60_xsub_keypoint_3d.py \
--work-dir work_dirs/2sagcn_80e_ntu60_xsub_keypoint_3d \
--validate --seed 0 --deterministic
Example: train AGCN model on bone data of NTU60 dataset in a deterministic option with periodic validation.
python tools/train.py configs/skeleton/2s-agcn/2sagcn_80e_ntu60_xsub_bone_3d.py \
--work-dir work_dirs/2sagcn_80e_ntu60_xsub_bone_3d \
--validate --seed 0 --deterministic
For more details, you can refer to Training setting part in getting_started.
Test
You can use the following command to test a model.
python tools/test.py ${CONFIG_FILE} ${CHECKPOINT_FILE} [optional arguments]
Example: test AGCN model on joint data of NTU60 dataset and dump the result to a pickle file.
python tools/test.py configs/skeleton/2s-agcn/2sagcn_80e_ntu60_xsub_keypoint_3d.py \
checkpoints/SOME_CHECKPOINT.pth --eval top_k_accuracy mean_class_accuracy \
--out joint_result.pkl
Example: test AGCN model on bone data of NTU60 dataset and dump the result to a pickle file.
python tools/test.py configs/skeleton/2s-agcn/2sagcn_80e_ntu60_xsub_bone_3d.py \
checkpoints/SOME_CHECKPOINT.pth --eval top_k_accuracy mean_class_accuracy \
--out bone_result.pkl
For more details, you can refer to Test a dataset part in getting_started.

 Datasets
Datasets
 Models
Models