|
a/README.md |
|
b/README.md |
| 1 |
# Multi-omics: state of the field |
1 |
# Multi-omics: state of the field |
| 2 |
|
2 |
|
| 3 |
[](https://travis-ci.com/krassowski/multi-omics-state-of-the-field) |
3 |
(https://travis-ci.com/krassowski/multi-omics-state-of-the-field)
|
| 4 |
[](https://mybinder.org/v2/gh/krassowski/multi-omics-state-of-the-field/HEAD?urlpath=lab/tree/notebooks) |
4 |
[](https://mybinder.org/v2/gh/krassowski/multi-omics-state-of-the-field/HEAD?urlpath=lab/tree/notebooks) |
| 5 |
|
5 |
|
| 6 |
Analyses for [State of the Field in Multi-Omics Research: From Computational Needs to Data Mining and Sharing](https://doi.org/10.3389/fgene.2020.610798). |
6 |
Analyses for [State of the Field in Multi-Omics Research: From Computational Needs to Data Mining and Sharing](https://doi.org/10.3389/fgene.2020.610798). |
| 7 |
|
7 |
|
| 8 |
## Overview |
8 |
## Overview |
| 9 |
|
9 |
|
| 10 |
[](https://github.com/krassowski/multi-omics-state-of-the-field/blob/master/figures/overview.pdf) |
10 |
[](https://github.com/krassowski/multi-omics-state-of-the-field/blob/master/figures/overview.pdf) |
| 11 |
|
11 |
|
| 12 |
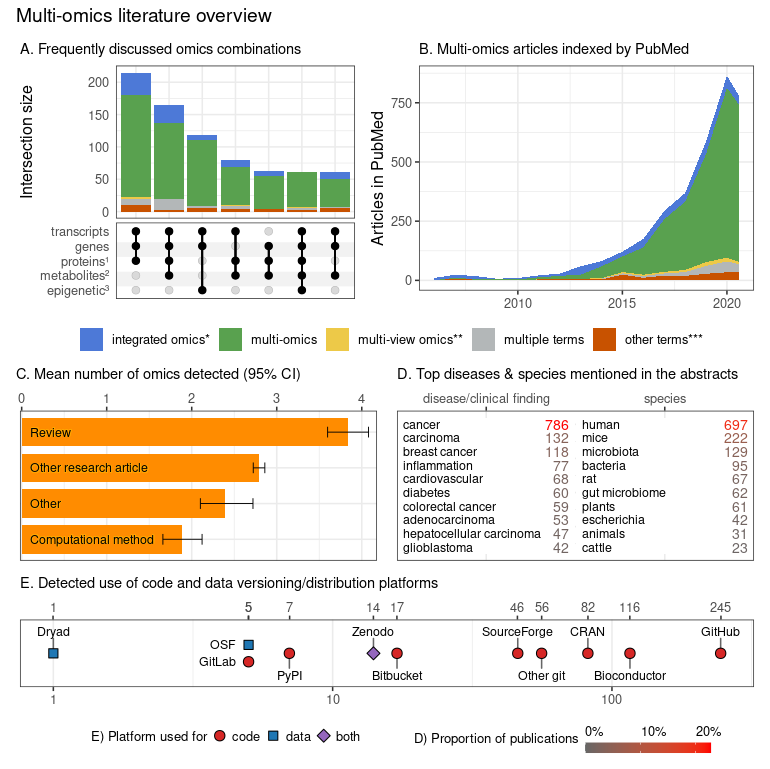
**Figure 1**. Characterization of multi-omics literature based on a systematic screen of PubMed indexed articles (up to July 2020). |
12 |
**Figure 1**. Characterization of multi-omics literature based on a systematic screen of PubMed indexed articles (up to July 2020). |
| 13 |
|
13 |
|
| 14 |
The comprehensive search terms (see the online repository for details) were collapsed into four categories; |
14 |
The comprehensive search terms (see the online repository for details) were collapsed into four categories;
|
| 15 |
_integrated omics_ (*) includes _integromics_ and _integrative_ omics, |
15 |
_integrated omics_ (*) includes _integromics_ and _integrative_ omics,
|
| 16 |
_multi-view_ (\*\*) includes multi-view|block|source|modal omics, |
16 |
_multi-view_ (\*\*) includes multi-view|block|source|modal omics,
|
| 17 |
_other terms_ (\*\*\*) include pan-, trans-, poly-, cross-omics. |
17 |
_other terms_ (\*\*\*) include pan-, trans-, poly-, cross-omics. |
| 18 |
|
18 |
|
| 19 |
The subpanels present: |
19 |
The subpanels present:
|
| 20 |
- A) Combinations of omics (grouped by the characterized entities) commonly discussed occurring together in multi-omics articles (intersections with ≥ 3 omics and at least 50 papers). |
20 |
- A) Combinations of omics (grouped by the characterized entities) commonly discussed occurring together in multi-omics articles (intersections with ≥ 3 omics and at least 50 papers).
|
| 21 |
The proteins group (1) also includes peptides; the metabolites group (2) includes other endogenous molecules; the epigenetic group (3) encompasses all epigenetic modifications. |
21 |
The proteins group (1) also includes peptides; the metabolites group (2) includes other endogenous molecules; the epigenetic group (3) encompasses all epigenetic modifications.
|
| 22 |
- B) Trend plot representing the rapidly increasing number of multi-omics articles indexed in PubMed (also after adjusting for the number of articles published in matched journals - data not shown); the dip in 2020 can be attributed to indexing delay which was not accounted for in the current plot. |
22 |
- B) Trend plot representing the rapidly increasing number of multi-omics articles indexed in PubMed (also after adjusting for the number of articles published in matched journals - data not shown); the dip in 2020 can be attributed to indexing delay which was not accounted for in the current plot.
|
| 23 |
- C) Distribution of articles categories that mention different numbers of omics; while it is understandable that multi-omics Reviews category discuss many omics, the Computational method category articles appear to lag behind all other article category types. |
23 |
- C) Distribution of articles categories that mention different numbers of omics; while it is understandable that multi-omics Reviews category discuss many omics, the Computational method category articles appear to lag behind all other article category types.
|
| 24 |
The detected number of omics may underestimate the actual numbers (due to the automated search strategy) but should put a useful lower bound on the number of omics discussed. |
24 |
The detected number of omics may underestimate the actual numbers (due to the automated search strategy) but should put a useful lower bound on the number of omics discussed.
|
| 25 |
Bootstrapped 95% confidence intervals around the mean are presented with the whiskers. |
25 |
Bootstrapped 95% confidence intervals around the mean are presented with the whiskers.
|
| 26 |
- D) The number of articles mentioning the most popular clinical findings, disease terms (here screening is based on ClinVar diseases list) and species (based upon NCBI Taxonomy database). |
26 |
- D) The number of articles mentioning the most popular clinical findings, disease terms (here screening is based on ClinVar diseases list) and species (based upon NCBI Taxonomy database).
|
| 27 |
Both databases were manually filtered down to remove ambiguous terms and merge plural/singular forms. |
27 |
Both databases were manually filtered down to remove ambiguous terms and merge plural/singular forms.
|
| 28 |
Only the abstracts were screened here. |
28 |
Only the abstracts were screened here.
|
| 29 |
- E) The detected references to code, data versioning, distribution platforms and systems (links to repositories with deposited code/data); both the abstracts and full-texts (open-access subset, 44% of all articles) were screened. |
29 |
- E) The detected references to code, data versioning, distribution platforms and systems (links to repositories with deposited code/data); both the abstracts and full-texts (open-access subset, 44% of all articles) were screened.
|
| 30 |
No manual curation to classify intent of the link inclusion (i.e. to share authors' code/data vs to report the use of a dataset/tool) was undertaken. |
30 |
No manual curation to classify intent of the link inclusion (i.e. to share authors' code/data vs to report the use of a dataset/tool) was undertaken. |
| 31 |
|
31 |
|
| 32 |
### Methods |
32 |
### Methods |
| 33 |
|
33 |
|
| 34 |
PubMed database was searched for articles pertaining to multi-omics on 25th July 2020, using fourteen terms (multi|pan|trans|poly|cross-omics, multi-table|source|view|modal|block omics, integrative omics, integrated omics and integromics) including plural/singular and hyphenated/unhyphenated variants combinations. |
34 |
PubMed database was searched for articles pertaining to multi-omics on 25th July 2020, using fourteen terms (multi|pan|trans|poly|cross-omics, multi-table|source|view|modal|block omics, integrative omics, integrated omics and integromics) including plural/singular and hyphenated/unhyphenated variants combinations.
|
| 35 |
The search was automated via Entrez E-utilities API and restricted to Text Words (to avoid matching articles based on the affiliation of authors to companies such as Panomics, Inc. or Integromics S.L.); the full text and additional metadata were retrieved from the PubMed Central (PMC) database for the open access subset of articles. |
35 |
The search was automated via Entrez E-utilities API and restricted to Text Words (to avoid matching articles based on the affiliation of authors to companies such as Panomics, Inc. or Integromics S.L.); the full text and additional metadata were retrieved from the PubMed Central (PMC) database for the open access subset of articles.
|
| 36 |
The feature extraction was performed via n-gram matching against ClinVar (diseases & clinical findings) and NCBI Taxonomy (species) databases, while omics references annotation was based on regular expressions capturing phrases with suffix -ome or -omic (accounting for multi-omic phrases and plural variants). |
36 |
The feature extraction was performed via n-gram matching against ClinVar (diseases & clinical findings) and NCBI Taxonomy (species) databases, while omics references annotation was based on regular expressions capturing phrases with suffix -ome or -omic (accounting for multi-omic phrases and plural variants).
|
| 37 |
All matches were manually filtered down to exclude false or irrelevant matches and to merge plural forms. |
37 |
All matches were manually filtered down to exclude false or irrelevant matches and to merge plural forms.
|
| 38 |
The article type was collated from five sources: |
38 |
The article type was collated from five sources:
|
| 39 |
- MeSH PublicationType as provided by PubMed, |
39 |
- MeSH PublicationType as provided by PubMed,
|
| 40 |
- community-maintained list of multi-omics software packages and methods: [mikelove/awesome-multi-omics](https://github.com/mikelove/awesome-multi-omics), |
40 |
- community-maintained list of multi-omics software packages and methods: [mikelove/awesome-multi-omics](https://github.com/mikelove/awesome-multi-omics),
|
| 41 |
- PMC-derived: |
41 |
- PMC-derived:
|
| 42 |
- ArticleType and |
42 |
- ArticleType and
|
| 43 |
- Subjects (journal-specific); |
43 |
- Subjects (journal-specific);
|
| 44 |
- manual annotation of articles published in Bioinformatics (Oxford, UK) due to lack of methods subject annotations in PMC data for this journal (performed by MK) |
44 |
- manual annotation of articles published in Bioinformatics (Oxford, UK) due to lack of methods subject annotations in PMC data for this journal (performed by MK) |
| 45 |
|
45 |
|
| 46 |
#### Flow diagram |
46 |
#### Flow diagram |
| 47 |
|
47 |
|
| 48 |
<img src="https://github.com/krassowski/multi-omics-state-of-the-field/blob/master/figures/flowchart.png?raw=true" title="Flowchart with counts" width=500> |
48 |
<img src="https://github.com/krassowski/multi-omics-state-of-the-field/blob/master/figures/flowchart.png?raw=true" title="Flowchart with counts" width=500> |
| 49 |
|
49 |
|
| 50 |
**Figure 2**. A flow diagram of the semi-automated multi-omics literature screening effort (up to July 2020). |
50 |
**Figure 2**. A flow diagram of the semi-automated multi-omics literature screening effort (up to July 2020). |
| 51 |
|
51 |
|
| 52 |
|
52 |
|
| 53 |
#### Code overview |
53 |
#### Code overview |
| 54 |
|
54 |
|
| 55 |
[](https://raw.githubusercontent.com/krassowski/multi-omics-state-of-the-field/master/figures/repository.svg) |
55 |
[](https://raw.githubusercontent.com/krassowski/multi-omics-state-of-the-field/master/figures/repository.svg) |
| 56 |
|
56 |
|
| 57 |
**Figure 3**. Overview of the notebooks in this code repository. Click on the plot to display an interactive version, from where you can open respective notebooks by clicking on the analysis nodes. |
57 |
**Figure 3**. Overview of the notebooks in this code repository. Click on the plot to display an interactive version, from where you can open respective notebooks by clicking on the analysis nodes. |
| 58 |
|
58 |
|
| 59 |
|
59 |
|
| 60 |
### Reference |
60 |
### Reference |
| 61 |
|
61 |
|
| 62 |
This analysis was contributed to our [introductory review of multi-omics field](https://doi.org/10.3389/fgene.2020.610798), now published in Frontiers in Genetics (open access): |
62 |
This analysis was contributed to our [introductory review of multi-omics field](https://doi.org/10.3389/fgene.2020.610798), now published in Frontiers in Genetics (open access): |
| 63 |
|
63 |
|
| 64 |
> Krassowski M, Das V, Sahu SK and Misra BB (2020) State of the Field in Multi-Omics Research: From Computational Needs to Data Mining and Sharing. Front. Genet. 11:610798. doi: 10.3389/fgene.2020.610798 |
64 |
Krassowski M, Das V, Sahu SK and Misra BB (2020) State of the Field in Multi-Omics Research: From Computational Needs to Data Mining and Sharing. Front. Genet. 11:610798. doi: 10.3389/fgene.2020.610798 |
| 65 |
|
65 |
|
| 66 |
|
66 |
|
| 67 |
### Reproducing |
67 |
### Reproducing |
| 68 |
|
68 |
|
| 69 |
Prerequisites: |
69 |
Prerequisites: |
| 70 |
|
70 |
|
| 71 |
- Ubuntu: 20.04 (x64) |
71 |
- Ubuntu: 20.04 (x64)
|
| 72 |
- Python: 3.8.3 |
72 |
- Python: 3.8.3
|
| 73 |
- R: 3.6.3 |
73 |
- R: 3.6.3 |
| 74 |
|
74 |
|
| 75 |
Install the minimal requirements for reproduction and download required data: |
75 |
Install the minimal requirements for reproduction and download required data: |
| 76 |
|
76 |
|
| 77 |
```bash |
77 |
```bash
|
| 78 |
pip install -r setup/requirements.txt |
78 |
pip install -r setup/requirements.txt
|
| 79 |
Rscript helpers/restore.R |
79 |
Rscript helpers/restore.R
|
| 80 |
cd data |
80 |
cd data
|
| 81 |
./download.sh |
81 |
./download.sh
|
| 82 |
``` |
82 |
``` |
| 83 |
|
83 |
|
| 84 |
|
84 |
|
| 85 |
### Development and contributing |
85 |
### Development and contributing |
| 86 |
|
86 |
|
| 87 |
Install additional requirements for development and testing: |
87 |
Install additional requirements for development and testing: |
| 88 |
|
88 |
|
| 89 |
```bash |
89 |
```bash
|
| 90 |
pip install -r setup/requirements-dev.txt |
90 |
pip install -r setup/requirements-dev.txt
|
| 91 |
``` |
91 |
``` |
| 92 |
|
92 |
|
| 93 |
Execute tests with: |
93 |
Execute tests with: |
| 94 |
|
94 |
|
| 95 |
```bash |
95 |
```bash
|
| 96 |
python3 -m pytest |
96 |
python3 -m pytest
|
| 97 |
``` |
97 |
``` |
| 98 |
|
98 |
|
| 99 |
Freeze (snapshot) R requirements with: |
99 |
Freeze (snapshot) R requirements with: |
| 100 |
|
100 |
|
| 101 |
```bash |
101 |
```bash
|
| 102 |
Rscript helpers/freeze.R |
102 |
Rscript helpers/freeze.R
|
| 103 |
``` |
103 |
``` |
| 104 |
|
104 |
|
| 105 |
Create the repository overview graph: |
105 |
Create the repository overview graph: |
| 106 |
|
106 |
|
| 107 |
```bash |
107 |
```bash
|
| 108 |
pip install nbpipeline |
108 |
pip install nbpipeline
|
| 109 |
PYTHONPATH=$(pwd):$PYTHONPATH nbpipeline --dry_run -s -O figures/repository.svg --display_graph_with none |
109 |
PYTHONPATH=$(pwd):$PYTHONPATH nbpipeline --dry_run -s -O figures/repository.svg --display_graph_with none
|
| 110 |
``` |
110 |
```
|