|
a |
|
b/README.md |
|
|
1 |
[](https://github.com/shenwanxiang/bidd-aggmap/tree/master/paper/example) |
|
|
2 |
[](https://bidd-aggmap.readthedocs.io/en/latest/?badge=latest) |
|
|
3 |
[](https://pepy.tech/project/aggmap) |
|
|
4 |
[](https://badge.fury.io/py/aggmap) |
|
|
5 |
[](https://academic.oup.com/nar/article/50/8/e45/6517966?login=false) |
|
|
6 |
|
|
|
7 |
|
|
|
8 |
<img src="./docs/images/logo.png" align="left" height="170" width="130" > |
|
|
9 |
|
|
|
10 |
|
|
|
11 |
|
|
|
12 |
# Jigsaw-like AggMap |
|
|
13 |
|
|
|
14 |
## A Robust and Explainable Omics Deep Learning Tool |
|
|
15 |
|
|
|
16 |
---- |
|
|
17 |
|
|
|
18 |
|
|
|
19 |
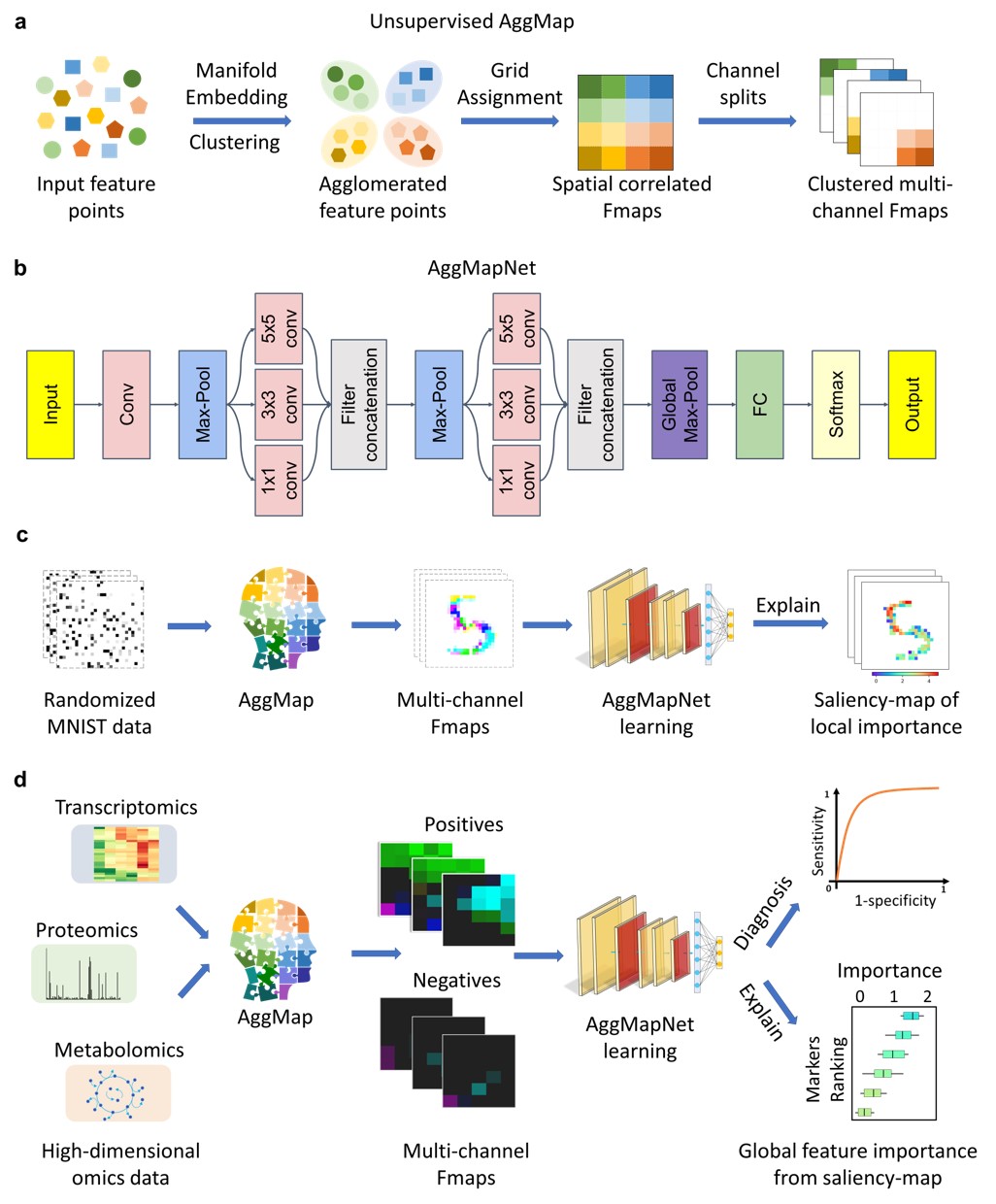
### Installation (Only on Linux system) |
|
|
20 |
|
|
|
21 |
install aggmap by: |
|
|
22 |
```bash |
|
|
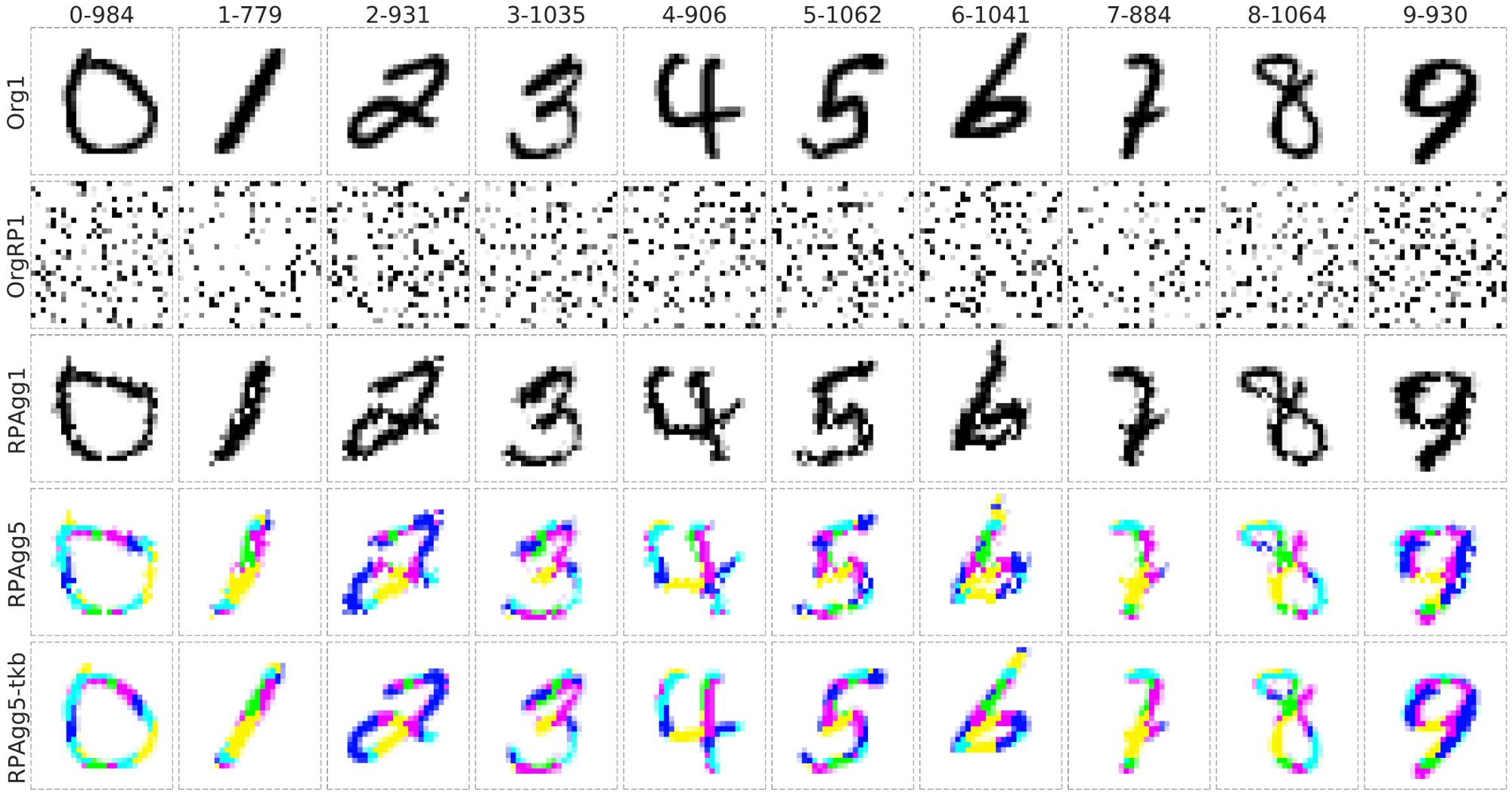
23 |
# create an aggmap env |
|
|
24 |
conda create -n aggmap python=3.8 |
|
|
25 |
conda activate aggmap |
|
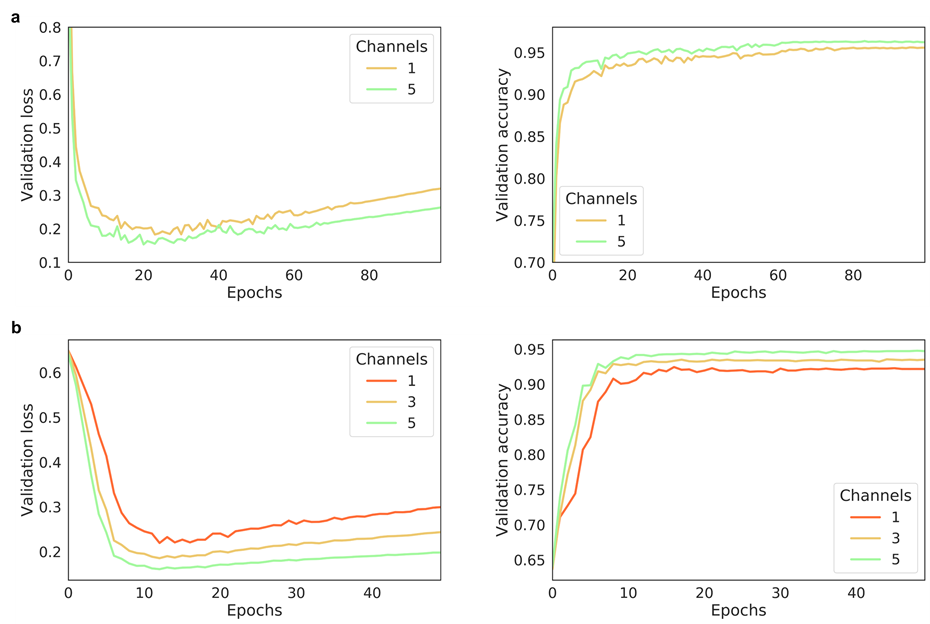
|
26 |
pip install --upgrade pip |
|
|
27 |
pip install aggmap==1.2.1 |
|
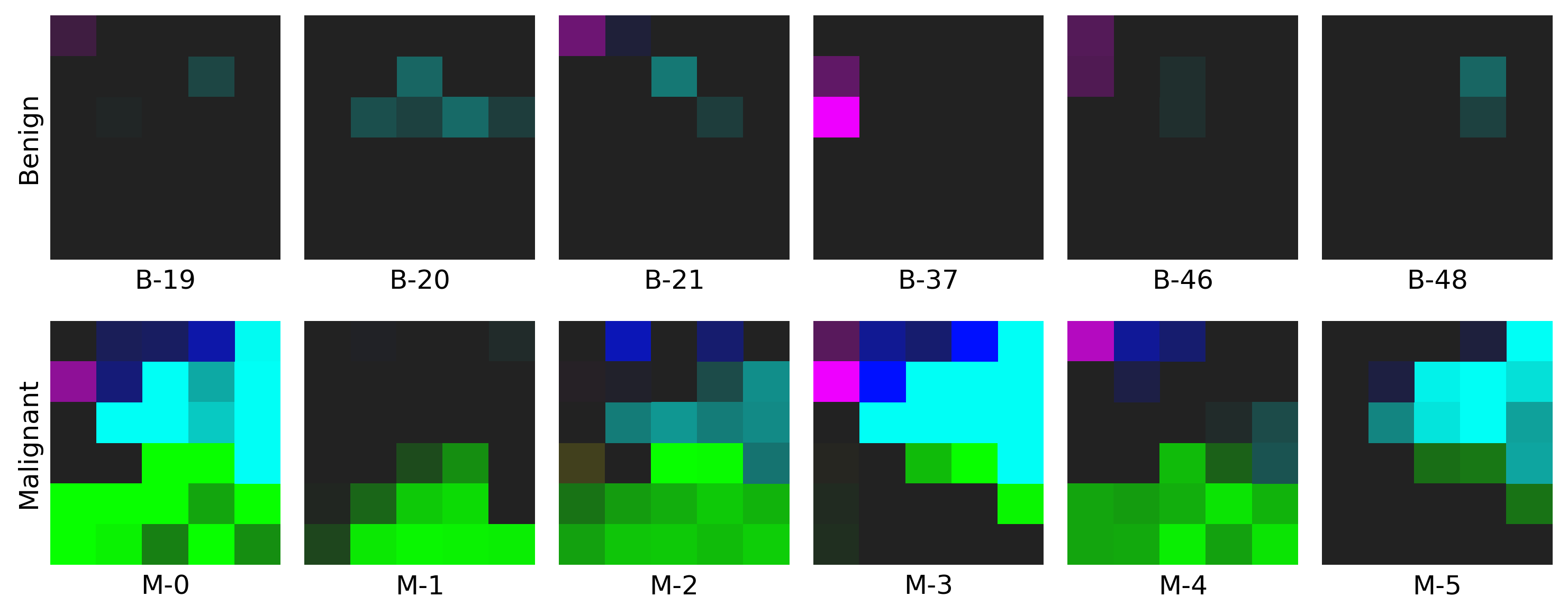
|
28 |
``` |
|
|
29 |
|
|
|
30 |
---- |
|
|
31 |
|
|
|
32 |
### Usage |
|
|
33 |
|
|
|
34 |
```python |
|
|
35 |
import pandas as pd |
|
|
36 |
from sklearn.datasets import load_breast_cancer |
|
|
37 |
from aggmap import AggMap, AggMapNet |
|
|
38 |
|
|
|
39 |
# Data loading |
|
|
40 |
data = load_breast_cancer() |
|
|
41 |
dfx = pd.DataFrame(data.data, columns=data.feature_names) |
|
|
42 |
dfy = pd.get_dummies(pd.Series(data.target)) |
|
|
43 |
|
|
|
44 |
# AggMap object definition, fitting, and saving |
|
|
45 |
mp = AggMap(dfx, metric = 'correlation') |
|
|
46 |
mp.fit(cluster_channels=5, emb_method = 'umap', verbose=0) |
|
|
47 |
mp.save('agg.mp') |
|
|
48 |
|
|
|
49 |
# AggMap visulizations: Hierarchical tree, embeddng scatter and grid |
|
|
50 |
mp.plot_tree() |
|
|
51 |
mp.plot_scatter(enabled_data_labels=True, radius=5) |
|
|
52 |
mp.plot_grid(enabled_data_labels=True) |
|
|
53 |
|
|
|
54 |
# Transoformation of 1d vectors to 3D Fmaps (-1, w, h, c) by AggMap |
|
|
55 |
X = mp.batch_transform(dfx.values, n_jobs=4, scale_method = 'minmax') |
|
|
56 |
y = dfy.values |
|
|
57 |
|
|
|
58 |
# AggMapNet training, validation, early stopping, and saving |
|
|
59 |
clf = AggMapNet.MultiClassEstimator(epochs=50, gpuid=0) |
|
|
60 |
clf.fit(X, y, X_valid=None, y_valid=None) |
|
|
61 |
clf.save_model('agg.model') |
|
|
62 |
|
|
|
63 |
# Model explaination by simply-explainer: global, local |
|
|
64 |
simp_explainer = AggMapNet.simply_explainer(clf, mp) |
|
|
65 |
global_simp_importance = simp_explainer.global_explain(clf.X_, clf.y_) |
|
|
66 |
local_simp_importance = simp_explainer.local_explain(clf.X_[[0]], clf.y_[[0]]) |
|
|
67 |
|
|
|
68 |
# Model explaination by shapley-explainer: global, local |
|
|
69 |
shap_explainer = AggMapNet.shapley_explainer(clf, mp) |
|
|
70 |
global_shap_importance = shap_explainer.global_explain(clf.X_) |
|
|
71 |
local_shap_importance = shap_explainer.local_explain(clf.X_[[0]]) |
|
|
72 |
``` |
|
|
73 |
|
|
|
74 |
|
|
|
75 |
### How It Works? |
|
|
76 |
|
|
|
77 |
- AggMap flowchart of feature mapping and agglomeration into ordered (spatially correlated) multi-channel feature maps (Fmaps) |
|
|
78 |
|
|
|
79 |
 |
|
|
80 |
**a**, AggMap flowchart of feature mapping and aggregation into ordered (spatially-correlated) channel-split feature maps (Fmaps).**b**, CNN-based AggMapNet architecture for Fmaps learning. **c**, proof-of-concept illustration of AggMap restructuring of unordered data (randomized MNIST) into clustered channel-split Fmaps (reconstructed MNIST) for CNN-based learning and important feature analysis. **d**, typical biomedical applications of AggMap in restructuring omics data into channel-split Fmaps for multi-channel CNN-based diagnosis and biomarker discovery (explanation `saliency-map` of important features). |
|
|
81 |
|
|
|
82 |
|
|
|
83 |
---- |
|
|
84 |
### Proof-of-Concepts of reconstruction ability on MNIST Dataset |
|
|
85 |
|
|
|
86 |
<video width="320" height="240" controls> |
|
|
87 |
<source src="https://www.shenwx.com/files/Video_MNIST.mp4" type="video/mp4"> |
|
|
88 |
</video> |
|
|
89 |
|
|
|
90 |
- It can reconstruct to the original image from completely randomly permuted (disrupted) MNIST data: |
|
|
91 |
|
|
|
92 |
|
|
|
93 |
|
|
|
94 |
 |
|
|
95 |
|
|
|
96 |
`Org1`: the original grayscale images (channel = 1), `OrgRP1`: the randomized images of Org1 (channel = 1), `RPAgg1, 5`: the reconstructed images of `OrgPR1` by AggMap feature restructuring (channel = 1, 5 respectively, each color represents features of one channel). `RPAgg5-tkb`: the original images with the pixels divided into 5 groups according to the 5-channels of `RPAgg5` and colored in the same way as `RPAgg5`. |
|
|
97 |
|
|
|
98 |
|
|
|
99 |
---- |
|
|
100 |
|
|
|
101 |
|
|
|
102 |
|
|
|
103 |
### The effect of the number of channels on model performance |
|
|
104 |
|
|
|
105 |
- Multi-channel Fmaps can boost the model performance notably: |
|
|
106 |
 |
|
|
107 |
|
|
|
108 |
The performance of AggMapNet using different number of channels on the `TCGA-T (a)` and `COV-D (b)`. For `TCGA-T`, ten-fold cross validation average performance, for `COV-D`, a fivefold cross validation was performed and repeat 5 rounds using different random seeds (total 25 training times), their average performances of the validation set were reported. |
|
|
109 |
---- |
|
|
110 |
|
|
|
111 |
|
|
|
112 |
### Example for Restructured Fmaps |
|
|
113 |
- The example on WDBC dataset: click [here](https://github.com/shenwanxiang/bidd-aggmap/blob/master/paper/example/00_breast_cancer/00_WDBC_example_flow.ipynb) to find out more! |
|
|
114 |
 |
|
|
115 |
|
|
|
116 |
---- |
|
|
117 |
|
|
|
118 |
|
|
|
119 |
|
|
|
120 |
### Citation |
|
|
121 |
Shen, Wan Xiang, et al. "AggMapNet: enhanced and explainable low-sample omics deep learning with feature-aggregated multi-channel networks." Nucleic Acids Research 50.8 (2022): e45-e45. |
|
|
122 |
|
|
|
123 |
---- |
|
|
124 |
|