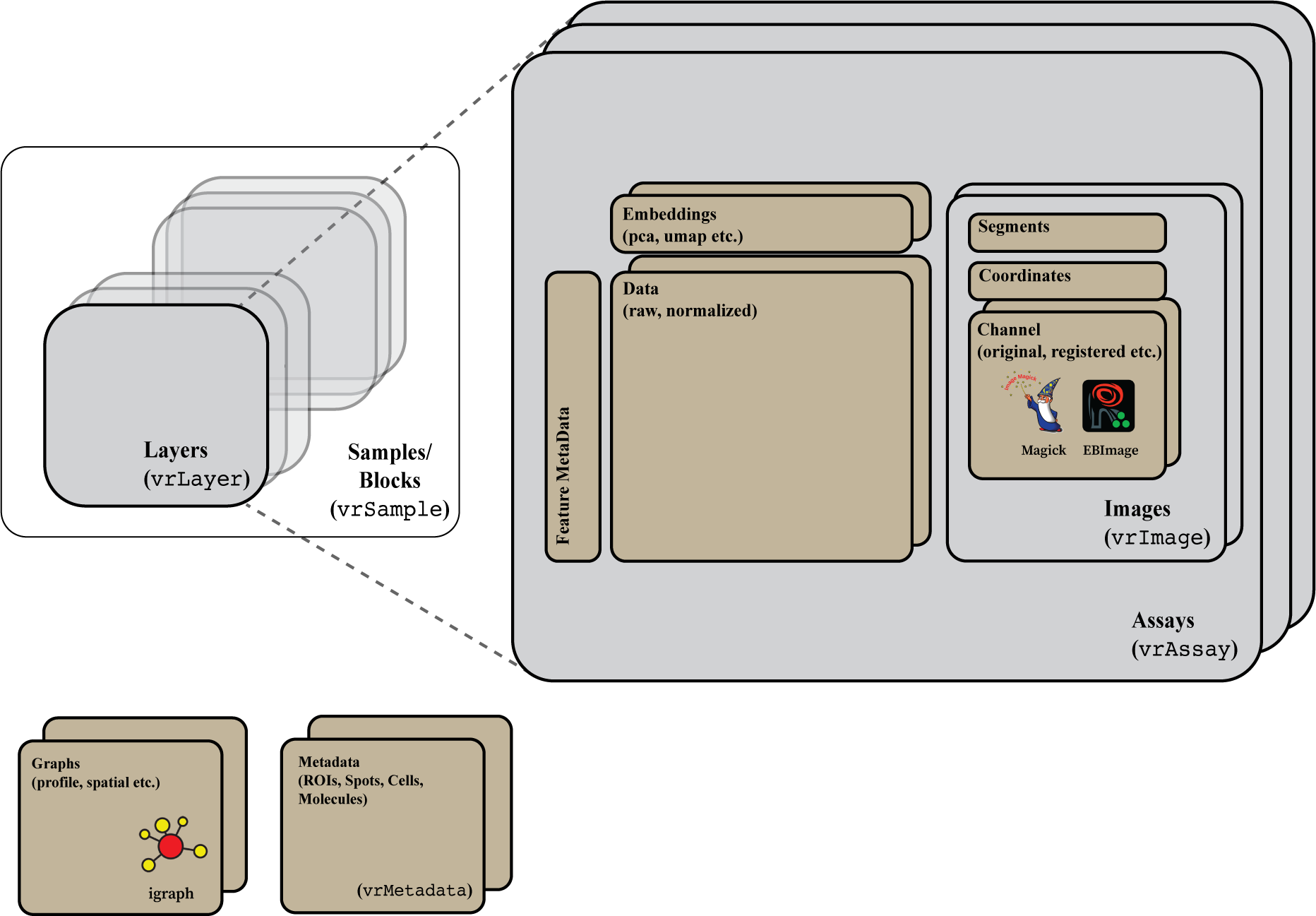
|
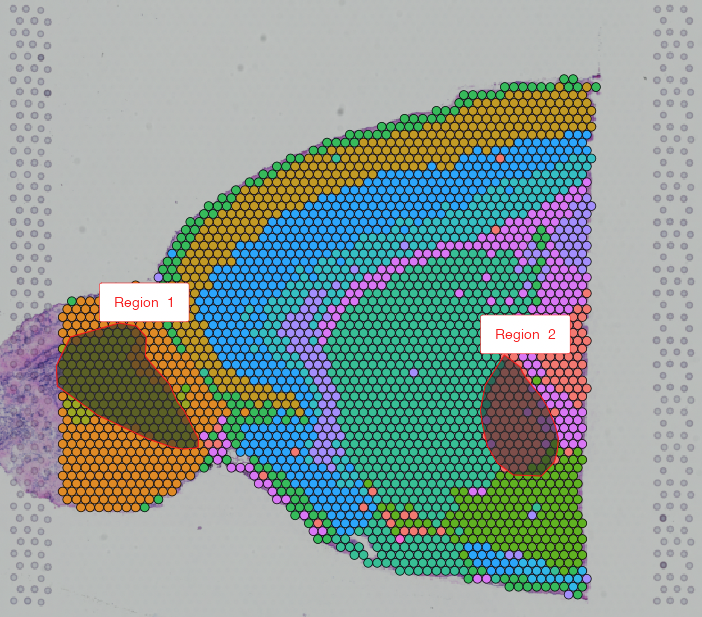
Spatial Data Alignment 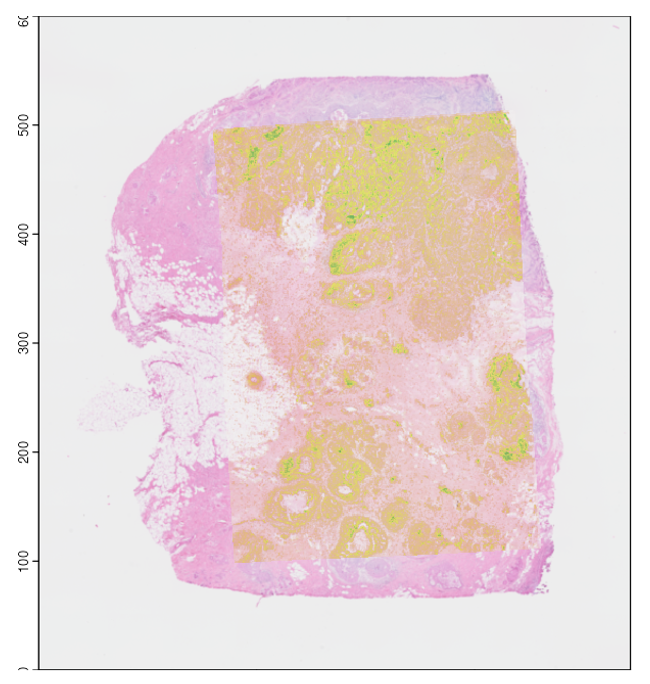 Automated and manual alignment of spatial data assays |
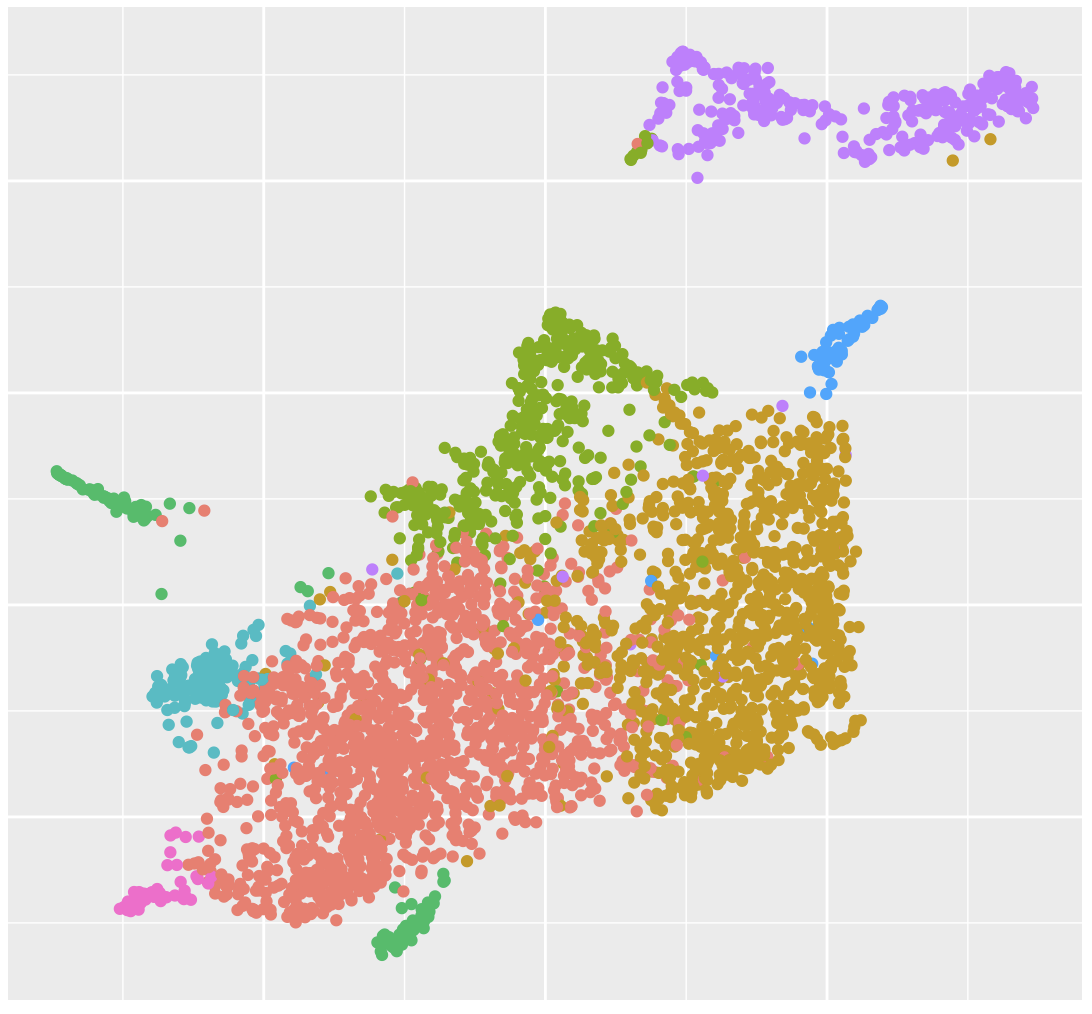
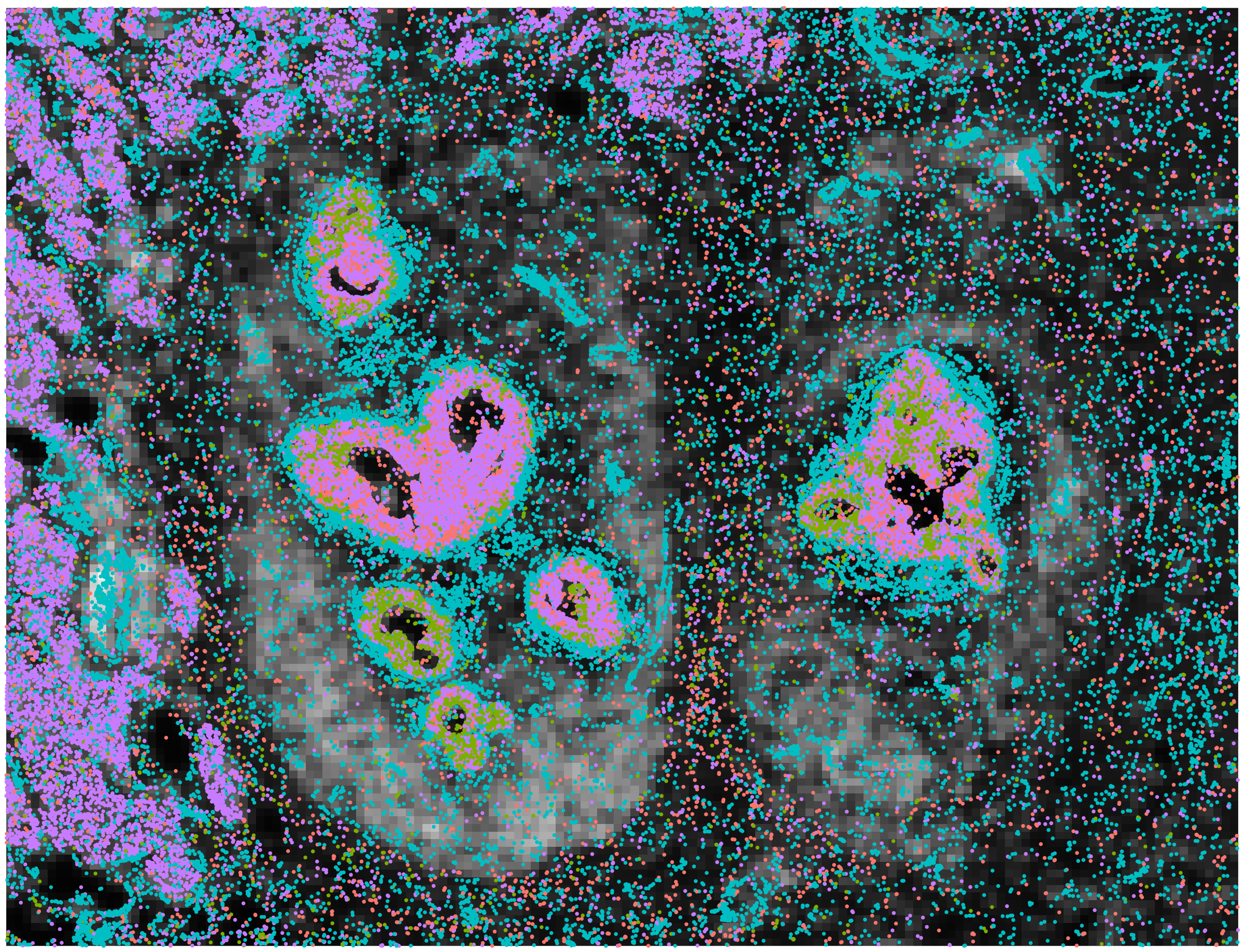
Multi-omic Integration 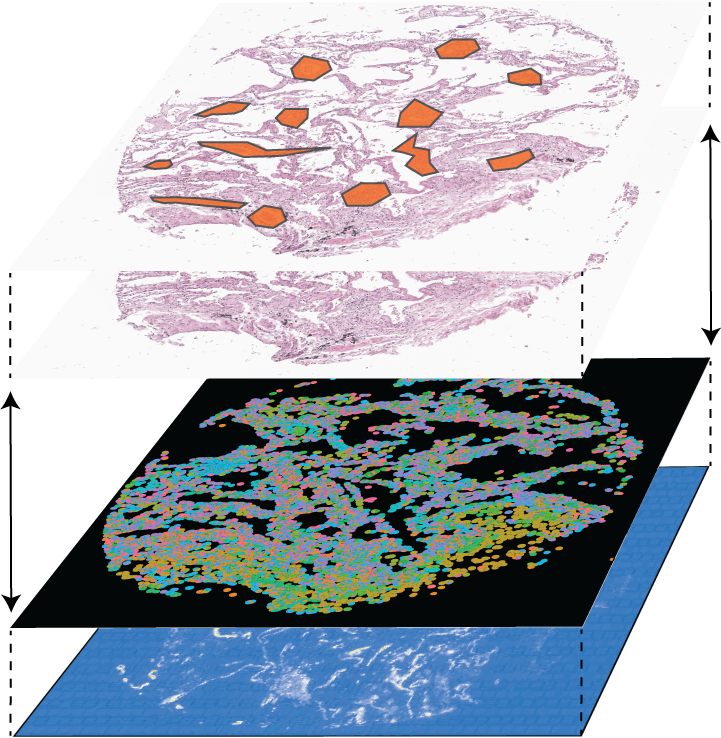 Integrating data modalities within or across tissue sections |
Niche Clustering 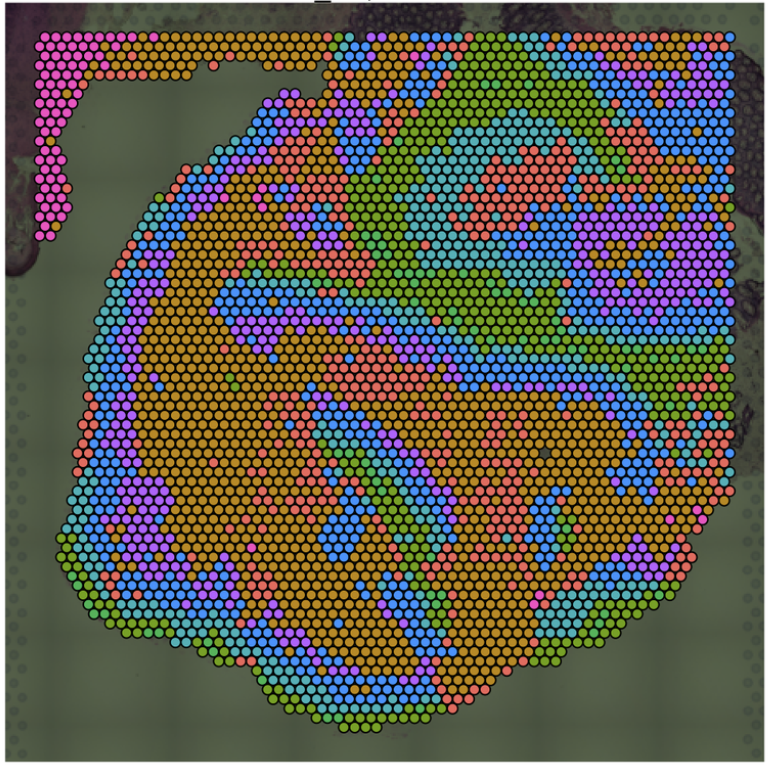 Clustering based on ROI/spot deconvolution and Spatial Neighborhood |