Diff of
/README.md
[000000]
..
[014e6e]
Switch to unified view
| a | b/README.md | ||
|---|---|---|---|
| 1 | |||
| 2 | ```shell |
||
| 3 | /$$$$$$ /$$ /$$$$$$$ /$$$$$$ |
||
| 4 | /$$__ $$ | $$ | $$__ $$ /$$__ $$ |
||
| 5 | | $$ \ $$ /$$ /$$ /$$$$$$ /$$$$$$ | $$ \ $$| $$ \ $$ |
||
| 6 | | $$$$$$$$| $$ | $$|_ $$_/ /$$__ $$| $$$$$$$ | $$$$$$$$ |
||
| 7 | | $$__ $$| $$ | $$ | $$ | $$ \ $$| $$__ $$| $$__ $$ |
||
| 8 | | $$ | $$| $$ | $$ | $$ /$$| $$ | $$| $$ \ $$| $$ | $$ |
||
| 9 | | $$ | $$| $$$$$$/ | $$$$/| $$$$$$/| $$$$$$$/| $$ | $$ |
||
| 10 | |__/ |__/ \______/ \___/ \______/ |_______/ |__/ |__/ |
||
| 11 | |||
| 12 | Automated Bioinformatics Analysis |
||
| 13 | www.joshuachou.ink/about |
||
| 14 | ``` |
||
| 15 | |||
| 16 | **An AI Agent for Fully Automated Multi-omic Analyses**. |
||
| 17 | |||
| 18 | (**Automated Bioinformatics Analysis via AutoBA**) |
||
| 19 | |||
| 20 | [Juexiao Zhou](https://www.joshuachou.ink/about/), Bin Zhang, Guowei Li, Xiuying Chen, Haoyang Li, Xiaopeng Xu, Siyuan Chen, Wenjia He, Chencheng Xu, Liwei Liu, Xin Gao |
||
| 21 | |||
| 22 | King Abdullah University of Science and Technology, KAUST |
||
| 23 | |||
| 24 | Huawei Technologies Co., Ltd |
||
| 25 | |||
| 26 | <a href='media/advs.202407094.pdf'><img src='https://img.shields.io/badge/Paper-PDF-red'></a> |
||
| 27 | |||
| 28 | https://github.com/JoshuaChou2018/AutoBA/assets/25849209/3334417a-de59-421c-aa5e-e2ac16ce90db |
||
| 29 | |||
| 30 | ## What's New |
||
| 31 | |||
| 32 | - **[2024/08]** Our paper is published online at [Advanced Science](https://onlinelibrary.wiley.com/doi/10.1002/advs.202407094) |
||
| 33 | - **[2024/08]** We integrated ollama to make it easier to use local LLMs and released the latest stable version `v0.4.0` |
||
| 34 | - **[2024/03]** Now we support retrieval-augmented generation (RAG) to increase robustness of AutoBA, to use it, please upgrade openai==1.13.3 and install llama-index. |
||
| 35 | - **[2024/02]** Now we support deepseek-coder-6.7b-instruct (failed test), deepseek-coder-7b-instruct-v1.5 (passed test), deepseek-coder-33b-instruct (passed test), to use it, please upgrade transformers==4.35.0. |
||
| 36 | - **[2024/01]** Don't like the command line mode? Now we provide a new GUI and released the milestone stable version `v0.2.0` 🎉 |
||
| 37 | - **[2024/01]** Updated JSON mode for gpt-3.5-turbo-1106, gpt-4-1106-preview, the output of these two models will be more stable |
||
| 38 | - **[2024/01]** Updated the support for ChatGPT-4 (gpt-4-32k-0613: Currently points to gpt-4-32k-0613, 32,768 tokens, Up to Sep 2021; gpt-4-1106-preview: GPT-4 Turbo, 128,000 tokens, Up to Apr 2023) |
||
| 39 | - **[2024/01]** Updated the support for ChatGPT-3.5 (gpt-3.5-turbo: openai chatgpt-3.5, 4,096 tokens and gpt-3.5-turbo-1106: openai chatgpt-3.5, 16,385 tokens) |
||
| 40 | - **[2023/12]** We added LLM support for the executor and the ACR module and released the milestone stable version `v0.1.1` |
||
| 41 | - **[2023/12]** We provided the latest docker version to simplify the installation process. |
||
| 42 | - **[2023/12]** New feature: automated code repairing (ACR module) added, add llama2-chat backends. |
||
| 43 | - **[2023/11]** We updated the executor and released the latest stable version (v0.0.2) and are working on automatic error feedback and code fixing. |
||
| 44 | - **[2023/09]** We integrated codellama 7b-Instruct, 13b-Instruct, 34b-Instruct, now users can choose to use chatgpt or local llm as backends, we currently recommend using chatgpt because tests have found that codellama is not as effective as chatgpt for complex bioinformatics tasks. |
||
| 45 | - **[2023/09]** We are pleased to announce the official release of AutoBA's latest version `v0.0.1`! 🎉🎉🎉 |
||
| 46 | |||
| 47 | ## TODO list |
||
| 48 | |||
| 49 | We're working hard to achieve more features, welcome to PRs! |
||
| 50 | |||
| 51 | - [x] Automatic error feedback and code fixing |
||
| 52 | - [x] Offer local LLMs (eg. code llama) as options for users |
||
| 53 | - [x] Provide a docker version, simplify the installation process |
||
| 54 | - [x] A UI-based YAML generator |
||
| 55 | - [x] Support deepseek coder |
||
| 56 | - [x] Support RAG |
||
| 57 | - [x] Support ollama |
||
| 58 | - [ ] Pack into a conda package, simplify the installation process |
||
| 59 | - [ ] Interactive mode |
||
| 60 | - [ ] GUI for data visualization |
||
| 61 | - [ ] Continue from breakpoint |
||
| 62 | - [ ] ... |
||
| 63 | - |
||
| 64 | **We appreciate all contributions to improve AutoBA.** |
||
| 65 | |||
| 66 | The `main` branch serves as the primary branch, while the development branch is `dev`. |
||
| 67 | |||
| 68 | Thank you for your unwavering support and enthusiasm, and let's work together to make AutoBA even more robust and powerful! If you want to contribute, please PR to the latest `dev` branch. 💪 |
||
| 69 | |||
| 70 | |||
| 71 | ## Installation |
||
| 72 | |||
| 73 | ### Command line |
||
| 74 | ```shell |
||
| 75 | # (mandatory) for basic functions |
||
| 76 | curl -L -O "https://github.com/conda-forge/miniforge/releases/latest/download/Mambaforge-$(uname)-$(uname -m).sh" |
||
| 77 | bash Mambaforge-$(uname)-$(uname -m).sh |
||
| 78 | git clone https://github.com/JoshuaChou2018/AutoBA.git |
||
| 79 | |||
| 80 | mamba create -n abc_runtime python==3.10 -y |
||
| 81 | mamba activate abc_runtime |
||
| 82 | # Then manually: |
||
| 83 | add conda-forge and bioconda to ~/mambaforge/.condarc |
||
| 84 | |||
| 85 | mamba create -n abc python==3.10 |
||
| 86 | mamba activate abc |
||
| 87 | mamba install -c anaconda yaml==0.2.5 -y |
||
| 88 | pip install openai==1.13.3 pyyaml==6.0 |
||
| 89 | pip install transformers==4.35.0 |
||
| 90 | pip install accelerate==0.29.2 |
||
| 91 | pip install bitsandbytes==0.43.1 |
||
| 92 | pip install vllm==0.4.1 |
||
| 93 | |||
| 94 | ## (optional) for RAG |
||
| 95 | pip install llama-index==0.10.14 |
||
| 96 | pip install llama-index-embeddings-huggingface |
||
| 97 | |||
| 98 | # (optional) for local llm with ollama |
||
| 99 | mamba install langchain-community==0.2.6 -y |
||
| 100 | curl -fsSL https://ollama.com/install.sh | OLLAMA_VERSION=0.3.4 sh |
||
| 101 | ## pull the model before using it with AutoBA |
||
| 102 | ollama run llama3.1 |
||
| 103 | |||
| 104 | # (optional) for gui version |
||
| 105 | pip install gradio==4.14.0 |
||
| 106 | |||
| 107 | # (optional) for local llm (llama2) |
||
| 108 | cd AutoBA/src/codellama-main |
||
| 109 | pip install -e . |
||
| 110 | |||
| 111 | ## apply for a download link at https://ai.meta.com/resources/models-and-libraries/llama-downloads/ |
||
| 112 | ## download codellama model weights: 7b-Instruct,13b-Instruct,34b-Instruct |
||
| 113 | cd src/codellama-main |
||
| 114 | bash download.sh |
||
| 115 | ## download llama2 model weights: 7B-chat,13B-chat,70B-chat |
||
| 116 | cd src/llama-main |
||
| 117 | bash download.sh |
||
| 118 | ## download hf version model weights |
||
| 119 | git lfs install |
||
| 120 | cd src/codellama-main |
||
| 121 | git clone https://huggingface.co/codellama/CodeLlama-7b-Instruct-hf |
||
| 122 | git clone https://huggingface.co/codellama/CodeLlama-13b-Instruct-hf |
||
| 123 | git clone https://huggingface.co/codellama/CodeLlama-34b-Instruct-hf |
||
| 124 | |||
| 125 | # (optional) for local llm (deepseek) |
||
| 126 | cd AutoBA/src/deepseek |
||
| 127 | git clone https://huggingface.co/deepseek-ai/deepseek-coder-6.7b-instruct |
||
| 128 | git clone https://huggingface.co/deepseek-ai/deepseek-coder-7b-instruct-v1.5 |
||
| 129 | git clone https://huggingface.co/deepseek-ai/deepseek-coder-33b-instruct |
||
| 130 | git clone https://huggingface.co/deepseek-ai/deepseek-llm-67b-chat |
||
| 131 | |||
| 132 | # (optional) for features under development: the yaml generator UI |
||
| 133 | pip install plotly==5.14.1 dash==2.9.3 pandas==2.0.1 dash-mantine-components==0.12.1 |
||
| 134 | ``` |
||
| 135 | |||
| 136 | ### Docker |
||
| 137 | |||
| 138 | Please refer to https://docs.docker.com/engine/install to install Docker first. |
||
| 139 | |||
| 140 | ```shell |
||
| 141 | # (mandatory) for basic functions |
||
| 142 | docker pull joshuachou666/autoba:cuda12.2.2-cudnn8-devel-ubuntu22.04-autoba0.1.2 |
||
| 143 | docker run --rm --gpus all -it joshuachou666/autoba:cuda12.2.2-cudnn8-devel-ubuntu22.04-autoba0.1.2 /bin/bash |
||
| 144 | ## Enter the shell in docker image |
||
| 145 | conda activate abc |
||
| 146 | cd AutoBA |
||
| 147 | ``` |
||
| 148 | |||
| 149 | If you get this error: **could not select device driver "" with capabilities: [[gpu]]**, then run the following codes: |
||
| 150 | ```shell |
||
| 151 | # (optional) for using GPU in docker |
||
| 152 | curl -s -L https://nvidia.github.io/nvidia-docker/gpgkey | \ |
||
| 153 | sudo apt-key add - |
||
| 154 | distribution=$(. /etc/os-release;echo $ID$VERSION_ID) |
||
| 155 | curl -s -L https://nvidia.github.io/nvidia-docker/$distribution/nvidia-docker.list | \ |
||
| 156 | sudo tee /etc/apt/sources.list.d/nvidia-docker.list |
||
| 157 | sudo apt-get update |
||
| 158 | sudo apt install -y nvidia-docker2 |
||
| 159 | sudo systemctl daemon-reload |
||
| 160 | sudo systemctl restart docker |
||
| 161 | ``` |
||
| 162 | |||
| 163 | Try the previous codes again. |
||
| 164 | |||
| 165 | ### Conda |
||
| 166 | ```shell |
||
| 167 | Coming soon... |
||
| 168 | ``` |
||
| 169 | |||
| 170 | ## Get Started |
||
| 171 | |||
| 172 | ### Understand files |
||
| 173 | |||
| 174 | `./example` contains several examples for you to start. |
||
| 175 | |||
| 176 | Under `./example`, `config.yaml` defines your files and goals. Defining `data_list`, `output_dir` and `goal_description` |
||
| 177 | in `config.yaml` is mandatory before running `app.py`. **Absolute paths rather than relative paths are recommended for all file paths defined in `config.yaml`**. |
||
| 178 | |||
| 179 | |||
| 180 | `app.py` run this file to start. |
||
| 181 | |||
| 182 | ### Start with one command |
||
| 183 | |||
| 184 | Run this command to start a simple example with chatgpt as backend (**recommended**). |
||
| 185 | |||
| 186 | `python app.py --config ./examples/case1.1/config.yaml --openai YOUR_OPENAI_API --model gpt-4` |
||
| 187 | |||
| 188 | Execute the code while generating it with ACR module loaded. |
||
| 189 | |||
| 190 | `python app.py --config ./examples/case1.1/config.yaml --openai YOUR_OPENAI_API --model gpt-4 --execute True` |
||
| 191 | |||
| 192 | **Please note that this work uses the GPT-4 API and does not guarantee that GPT-3.5 will work properly in all cases.** |
||
| 193 | |||
| 194 | or with local llm as backend (**not recommended for the moment, in development and only for testing purposes**) |
||
| 195 | |||
| 196 | `python app.py --config ./examples/case1.1/config.yaml --model codellama-7bi` |
||
| 197 | |||
| 198 | or with local llm based on ollama as backend |
||
| 199 | |||
| 200 | `python app.py --config ./examples/case1.1/config.yaml --model ollama_llama3.1` |
||
| 201 | |||
| 202 | ### Start GUI version |
||
| 203 | |||
| 204 | Run this command to start a GUI version of AutoBA. |
||
| 205 | |||
| 206 | `python gui.py` |
||
| 207 | |||
| 208 | 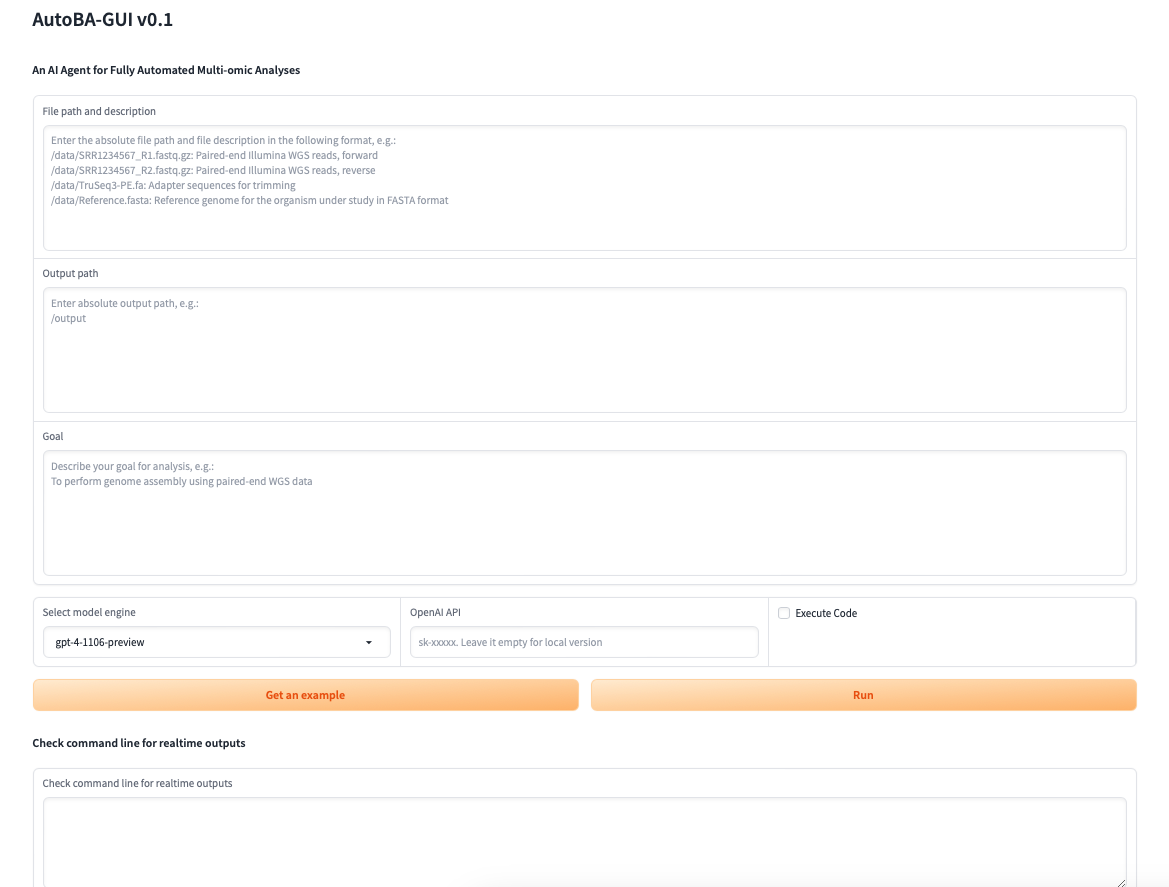 |
||
| 209 | |||
| 210 | ### Model Zoo |
||
| 211 | |||
| 212 | **Dynamic Engine: dynamic update version** |
||
| 213 | - gpt-3.5-turbo: Points to the latest gpt-3.5 model |
||
| 214 | - gpt-4-turbo: Points to the latest gpt-4 model |
||
| 215 | - gpt-4o: Points to the latest gpt-4o model |
||
| 216 | - gpt-4o-mini: Points to the latest gpt-4o-mini model |
||
| 217 | - gpt-4: (default) |
||
| 218 | - For more information, please check: https://platform.openai.com/docs/models/gpt-4-turbo-and-gpt-4 |
||
| 219 | |||
| 220 | **Ollama Engine:** |
||
| 221 | - ollama_llama3.1: llama3.1 |
||
| 222 | - ollama_llama3.1:8b: llama3.1:8b |
||
| 223 | - ollama_mistral: mistral |
||
| 224 | - ... |
||
| 225 | - the `ollama_` prefix is mandatory, for more models, please refer to https://ollama.com/library |
||
| 226 | |||
| 227 | **Fixed Engine: snapshot version** |
||
| 228 | - gpt-3.5-turbo-1106: Updated GPT 3.5 Turbo, 16,385 tokens, Up to Sep 2021 |
||
| 229 | - gpt-4-0613: Snapshot of gpt-4 from June 13th 2023 with improved function calling support, 8,192 tokens, Up to Sep 2021 |
||
| 230 | - gpt-4-32k-0613: Snapshot of gpt-4-32k from June 13th 2023 with improved function calling support, 32,768 tokens, Up to Sep 2021 |
||
| 231 | - gpt-4-1106-preview: GPT-4 Turbo, 128,000 tokens, Up to Apr 2023 |
||
| 232 | - codellama-7bi: 7b-Instruct |
||
| 233 | - codellama-13bi: 13b-Instruct |
||
| 234 | - codellama-34bi: 34b-Instruct |
||
| 235 | - llama2-7bc: llama-2-7b-chat |
||
| 236 | - llama2-13bc: llama-2-13b-chat |
||
| 237 | - llama2-70bc: llama-2-70b-chat |
||
| 238 | - deepseek-6.7bi: deepseek-coder-6.7b-instruct |
||
| 239 | - deepseek-7bi: deepseek-coder-7b-instruct-v1.5 |
||
| 240 | - deepseek-33bi: deepseek-coder-33b-instruct |
||
| 241 | - deepseek-67bc: deepseek-llm-67b-chat |
||
| 242 | |||
| 243 | ## Use Cases |
||
| 244 | |||
| 245 | These cases below may have different ID numbers as those cases in our paper. |
||
| 246 | |||
| 247 | ### Example 1: Bulk RNA-Seq |
||
| 248 | |||
| 249 | #### Case 1.1: Find differentially expressed genes |
||
| 250 | |||
| 251 | **Reference**: https://pzweuj.github.io/worstpractice/site/C02_RNA-seq/01.prepare_data/ |
||
| 252 | |||
| 253 | Design of `config.yaml` |
||
| 254 | ```yaml |
||
| 255 | data_list: [ './examples/case1.1/data/SRR1374921.fastq.gz: single-end mouse rna-seq reads, replicate 1 in LoGlu group', |
||
| 256 | './examples/case1.1/data/SRR1374922.fastq.gz: single-end mouse rna-seq reads, replicate 2 in LoGlu group', |
||
| 257 | './examples/case1.1/data/SRR1374923.fastq.gz: single-end mouse rna-seq reads, replicate 1 in HiGlu group', |
||
| 258 | './examples/case1.1/data/SRR1374924.fastq.gz: single-end mouse rna-seq reads, replicate 2 in HiGlu group', |
||
| 259 | './examples/case1.1/data/TruSeq3-SE.fa: trimming adapter', |
||
| 260 | './examples/case1.1/data/mm39.fa: mouse mm39 genome fasta', |
||
| 261 | './examples/case1.1/data/mm39.ncbiRefSeq.gtf: mouse mm39 genome annotation' ] |
||
| 262 | output_dir: './examples/case1.1/output' |
||
| 263 | goal_description: 'find the differentially expressed genes' |
||
| 264 | ``` |
||
| 265 | |||
| 266 | ##### Download Data |
||
| 267 | ```shell |
||
| 268 | wget -P data/ http://hgdownload.soe.ucsc.edu/goldenPath/mm39/bigZips/genes/mm39.ncbiRefSeq.gtf.gz |
||
| 269 | wget -P data/ http://hgdownload.soe.ucsc.edu/goldenPath/mm39/bigZips/mm39.fa.gz |
||
| 270 | gunzip data/mm39.ncbiRefSeq.gtf.gz |
||
| 271 | gunzip data/mm39.fa.gz |
||
| 272 | wget -P data/ ftp://ftp.sra.ebi.ac.uk/vol1/fastq/SRR137/001/SRR1374921/SRR1374921.fastq.gz |
||
| 273 | wget -P data/ ftp://ftp.sra.ebi.ac.uk/vol1/fastq/SRR137/002/SRR1374922/SRR1374922.fastq.gz |
||
| 274 | wget -P data/ ftp://ftp.sra.ebi.ac.uk/vol1/fastq/SRR137/003/SRR1374923/SRR1374923.fastq.gz |
||
| 275 | wget -P data/ ftp://ftp.sra.ebi.ac.uk/vol1/fastq/SRR137/004/SRR1374924/SRR1374924.fastq.gz |
||
| 276 | ``` |
||
| 277 | |||
| 278 | ##### Analyze with AutoBA |
||
| 279 | |||
| 280 | ```shell |
||
| 281 | python app.py --config ./examples/case1.1/config.yaml --openai YOUR_OPENAI_API --model gpt-4 |
||
| 282 | python app.py --config ./examples/case1.1/config.yaml --model codellama-7bi |
||
| 283 | python app.py --config ./examples/case1.1/config.yaml --model codellama-13bi |
||
| 284 | python app.py --config ./examples/case1.1/config.yaml --model codellama-34bi |
||
| 285 | ``` |
||
| 286 | |||
| 287 | #### Case 1.2: Identify top5 down-regulated genes in HiGlu group |
||
| 288 | |||
| 289 | **Reference**: https://pzweuj.github.io/worstpractice/site/C02_RNA-seq/01.prepare_data/ |
||
| 290 | |||
| 291 | Design of `config.yaml` |
||
| 292 | ```yaml |
||
| 293 | data_list: [ './examples/case1.2/data/SRR1374921.fastq.gz: single-end mouse rna-seq reads, replicate 1 in LoGlu group', |
||
| 294 | './examples/case1.2/data/SRR1374922.fastq.gz: single-end mouse rna-seq reads, replicate 2 in LoGlu group', |
||
| 295 | './examples/case1.2/data/SRR1374923.fastq.gz: single-end mouse rna-seq reads, replicate 1 in HiGlu group', |
||
| 296 | './examples/case1.2/data/SRR1374924.fastq.gz: single-end mouse rna-seq reads, replicate 2 in HiGlu group', |
||
| 297 | './examples/case1.2/data/TruSeq3-SE.fa: trimming adapter', |
||
| 298 | './examples/case1.2/data/mm39.fa: mouse mm39 genome fasta', |
||
| 299 | './examples/case1.2/data/mm39.ncbiRefSeq.gtf: mouse mm39 genome annotation' ] |
||
| 300 | output_dir: './examples/case1.2/output' |
||
| 301 | goal_description: 'Identify top5 down-regulated genes in HiGlu group' |
||
| 302 | ``` |
||
| 303 | |||
| 304 | ##### Download Data |
||
| 305 | ```shell |
||
| 306 | wget -P data/ http://hgdownload.soe.ucsc.edu/goldenPath/mm39/bigZips/genes/mm39.ncbiRefSeq.gtf.gz |
||
| 307 | wget -P data/ http://hgdownload.soe.ucsc.edu/goldenPath/mm39/bigZips/mm39.fa.gz |
||
| 308 | gunzip data/mm39.ncbiRefSeq.gtf.gz |
||
| 309 | gunzip data/mm39.fa.gz |
||
| 310 | wget -P data/ ftp://ftp.sra.ebi.ac.uk/vol1/fastq/SRR137/001/SRR1374921/SRR1374921.fastq.gz |
||
| 311 | wget -P data/ ftp://ftp.sra.ebi.ac.uk/vol1/fastq/SRR137/002/SRR1374922/SRR1374922.fastq.gz |
||
| 312 | wget -P data/ ftp://ftp.sra.ebi.ac.uk/vol1/fastq/SRR137/003/SRR1374923/SRR1374923.fastq.gz |
||
| 313 | wget -P data/ ftp://ftp.sra.ebi.ac.uk/vol1/fastq/SRR137/004/SRR1374924/SRR1374924.fastq.gz |
||
| 314 | ``` |
||
| 315 | |||
| 316 | ```shell |
||
| 317 | python app.py --config ./examples/case1.2/config.yaml |
||
| 318 | ``` |
||
| 319 | |||
| 320 | #### Case 1.3: Predict Fusion genes using STAR-Fusion |
||
| 321 | |||
| 322 | **Reference:** https://github.com/STAR-Fusion/STAR-Fusion-Tutorial/wiki |
||
| 323 | |||
| 324 | Design of `config.yaml` |
||
| 325 | ```yaml |
||
| 326 | data_list: [ './examples/case1.3/data/rnaseq_1.fastq.gz: RNA-Seq read 1 data (left read)', |
||
| 327 | './examples/case1.3/data/rnaseq_2.fastq.gz: RNA-Seq read 2 data (right read)', |
||
| 328 | './examples/case1.3/data/CTAT_HumanFusionLib.mini.dat.gz: a small fusion annotation library', |
||
| 329 | './examples/case1.3/data/minigenome.fa: small genome sequence consisting of ~750 genes.', |
||
| 330 | './examples/case1.3/data/minigenome.gtf: transcript structure annotations for these genes' ] |
||
| 331 | output_dir: './examples/case1.3/output' |
||
| 332 | goal_description: 'Predict Fusion genes using STAR-Fusion' |
||
| 333 | ``` |
||
| 334 | |||
| 335 | ##### Download Data |
||
| 336 | |||
| 337 | ```shell |
||
| 338 | cd data |
||
| 339 | git clone https://github.com/STAR-Fusion/STAR-Fusion-Tutorial.git |
||
| 340 | mv STAR-Fusion-Tutorial/* . |
||
| 341 | ``` |
||
| 342 | |||
| 343 | ### Example 2: Single Cell RNA-seq Analysis |
||
| 344 | |||
| 345 | #### Case 2.1: Find the differentially expressed genes |
||
| 346 | |||
| 347 | **Reference:** https://www.jianshu.com/p/e22a947e6c60 |
||
| 348 | |||
| 349 | Design of `config.yaml` |
||
| 350 | ```yaml |
||
| 351 | data_list: [ './examples/case2.1/data/filtered_gene_bc_matrices/hg19: path to 10x mtx data',] |
||
| 352 | output_dir: './examples/case2.1/output' |
||
| 353 | goal_description: 'use scanpy to find the differentially expressed genes' |
||
| 354 | ``` |
||
| 355 | |||
| 356 | ##### Download Data |
||
| 357 | |||
| 358 | ```shell |
||
| 359 | wget -P data/ http://cf.10xgenomics.com/samples/cell-exp/1.1.0/pbmc3k/pbmc3k_filtered_gene_bc_matrices.tar.gz |
||
| 360 | cd data |
||
| 361 | tar zxvf pbmc3k_filtered_gene_bc_matrices.tar.gz |
||
| 362 | ``` |
||
| 363 | |||
| 364 | #### Case 2.2: Perform clustering |
||
| 365 | |||
| 366 | **Reference:** https://www.jianshu.com/p/e22a947e6c60 |
||
| 367 | |||
| 368 | Design of `config.yaml` |
||
| 369 | ```yaml |
||
| 370 | data_list: [ './examples/case2.2/data/filtered_gene_bc_matrices/hg19: path to 10x mtx data',] |
||
| 371 | output_dir: './examples/case2.2/output' |
||
| 372 | goal_description: 'use scanpy to perform clustering and visualize the expression level of gene PPBP in the UMAP.' |
||
| 373 | ``` |
||
| 374 | |||
| 375 | #### Case 2.3: Identify top5 marker genes |
||
| 376 | |||
| 377 | **Reference:** https://www.jianshu.com/p/e22a947e6c60 |
||
| 378 | |||
| 379 | Design of `config.yaml` |
||
| 380 | ```yaml |
||
| 381 | data_list: [ './examples/case2.3/data/filtered_gene_bc_matrices/hg19: path to 10x mtx data',] |
||
| 382 | output_dir: './examples/case2.3/output' |
||
| 383 | goal_description: 'use scanpy to identify top5 marker genes' |
||
| 384 | ``` |
||
| 385 | |||
| 386 | ### Example 3: ChIP-Seq Analysis |
||
| 387 | |||
| 388 | #### Case 3.1: Call peaks |
||
| 389 | |||
| 390 | **Reference:** https://pzweuj.github.io/2018/08/22/chip-seq-workflow.html |
||
| 391 | |||
| 392 | Design of `config.yaml` |
||
| 393 | |||
| 394 | |||
| 395 | ##### Download Data |
||
| 396 | |||
| 397 | ```shell |
||
| 398 | # 下载了5个样本,分别是Ring1B、cbx7、SUZ12、RYBP、IgGold。 |
||
| 399 | for ((i=204; i<=208; i++)); \ |
||
| 400 | do wget ftp://ftp.sra.ebi.ac.uk/vol1/fastq/SRR620/SRR620$i/SRR620$i.fastq.gz; \ |
||
| 401 | done |
||
| 402 | |||
| 403 | wget http://hgdownload.soe.ucsc.edu/goldenPath/mm39/bigZips/genes/mm39.ncbiRefSeq.gtf.gz |
||
| 404 | wget http://hgdownload.soe.ucsc.edu/goldenPath/mm39/bigZips/mm39.fa.gz |
||
| 405 | gunzip mm39.ncbiRefSeq.gtf.gz |
||
| 406 | gunzip mm39.fa.gz |
||
| 407 | ``` |
||
| 408 | |||
| 409 | Design of `config.yaml` |
||
| 410 | ```yaml |
||
| 411 | data_list: [ './examples/case3.1/data/SRR620204.fastq.gz: chip-seq data for Ring1B', |
||
| 412 | './examples/case3.1/data/SRR620205.fastq.gz: chip-seq data for cbx7', |
||
| 413 | './examples/case3.1/data/SRR620206.fastq.gz: chip-seq data for SUZ12', |
||
| 414 | './examples/case3.1/data/SRR620208.fastq.gz: chip-seq data for IgGold', |
||
| 415 | './examples/case3.1/data/mm39.ncbiRefSeq.gtf: genome annotations mouse', |
||
| 416 | './examples/case3.1/data/mm39.fa: mouse genome' ] |
||
| 417 | output_dir: './examples/case3.1/output' |
||
| 418 | goal_description: 'call peaks for protein cbx7 with IgGold as control' |
||
| 419 | ``` |
||
| 420 | |||
| 421 | #### Case 3.2: Discover motifs within the peaks |
||
| 422 | |||
| 423 | Design of `config.yaml` |
||
| 424 | ```yaml |
||
| 425 | data_list: [ './examples/case3.2/data/SRR620204.fastq.gz: chip-seq data for Ring1B', |
||
| 426 | './examples/case3.2/data/SRR620205.fastq.gz: chip-seq data for cbx7', |
||
| 427 | './examples/case3.2/data/SRR620206.fastq.gz: chip-seq data for SUZ12', |
||
| 428 | './examples/case3.2/data/SRR620208.fastq.gz: chip-seq data for IgGold', |
||
| 429 | './examples/case3.2/data/mm39.ncbiRefSeq.gtf: genome annotations mouse', |
||
| 430 | './examples/case3.2/data/mm39.fa: mouse genome' ] |
||
| 431 | output_dir: './examples/case3.2/output' |
||
| 432 | goal_description: 'Discover motifs within the peaks of protein SUZ12 with IgGold as control' |
||
| 433 | ``` |
||
| 434 | |||
| 435 | #### Case 3.3: Functional Enrichment |
||
| 436 | Design of `config.yaml` |
||
| 437 | ```yaml |
||
| 438 | data_list: [ './examples/case3.3/data/SRR620204.fastq.gz: chip-seq data for Ring1B', |
||
| 439 | './examples/case3.3/data/SRR620205.fastq.gz: chip-seq data for cbx7', |
||
| 440 | './examples/case3.3/data/SRR620206.fastq.gz: chip-seq data for SUZ12', |
||
| 441 | './examples/case3.3/data/SRR620208.fastq.gz: chip-seq data for IgGold', |
||
| 442 | './examples/case3.3/data/mm39.ncbiRefSeq.gtf: genome annotations mouse', |
||
| 443 | './examples/case3.3/data/mm39.fa: mouse genome' ] |
||
| 444 | output_dir: './examples/case3.3/output' |
||
| 445 | goal_description: 'perform functional enrichment for protein Ring1B, use protein IgGold as the control' |
||
| 446 | ``` |
||
| 447 | |||
| 448 | ### Example 4: Spatial Transcriptomics |
||
| 449 | |||
| 450 | #### Case 4.1: Neighborhood enrichment analysis |
||
| 451 | |||
| 452 | **Reference:** https://squidpy.readthedocs.io/en/latest/notebooks/tutorials/tutorial_seqfish.html |
||
| 453 | |||
| 454 | Design of `config.yaml` |
||
| 455 | ```yaml |
||
| 456 | data_list: [ './examples/case4.1/data/slice1.h5ad: spatial transcriptomics data for slice 1 in AnnData format',] |
||
| 457 | output_dir: './examples/case4.1/output' |
||
| 458 | goal_description: 'use squidpy for neighborhood enrichment analysis' |
||
| 459 | ``` |
||
| 460 | |||
| 461 | ## Custom Examples for Users |
||
| 462 | |||
| 463 | To use AutoBA in your case, please copy `config.yaml` to your destination and modify it accordingly. |
||
| 464 | Then you are ready to go. We welcome all developers to submit PR to upload your special cases under `./projects` |
||
| 465 | |||
| 466 | ## Citation |
||
| 467 | |||
| 468 | If you find this project useful in your research, please consider citing: |
||
| 469 | |||
| 470 | J. Zhou, B. Zhang, G. Li, X. Chen, H. Li, X. Xu, S. Chen, W. He, C. Xu, L. Liu, X. Gao, An AI Agent for Fully Automated Multi-Omic Analyses. Adv. Sci. 2024, 2407094. https://doi.org/10.1002/advs.202407094 |
||
| 471 | |||
| 472 | ## License |
||
| 473 | |||
| 474 | This project is released under the MIT license. |

 Datasets
Datasets
 Models
Models